
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,907,610 – 14,907,714 |
| Length | 104 |
| Max. P | 0.848683 |

| Location | 14,907,610 – 14,907,714 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.74 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -17.16 |
| Energy contribution | -17.28 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14907610 104 + 27905053 AUGGCCAAGUGGGGAUCUGCUUGCCUCCCAGCU-CACAUACCUGUGGUGGUGGCGGCGCGUCUUAUUAGAAUUUAUGA-UUCCCAUCU---------UUUUCUUUUUUGCUUUCU ..((..(((((((((((.(((((((.((..((.-((((....)))))))).))))).)).(((....)))......))-)))))).))---------)..))............. ( -28.80) >DroSim_CAF1 34896 113 + 1 AUGGCCAAGUGGGGAUCUGCUUGCCUCCCAGCU-CACAUACCUGUGGUGGUGGCGGCGCGUCUUAUUAGAAUUUAUGA-UUCCCAUCUAUUUUUUCUUUUUUUUUUUUGCUUUCU ........(((((((((.(((((((.((..((.-((((....)))))))).))))).)).(((....)))......))-)))))))............................. ( -28.50) >DroEre_CAF1 34363 94 + 1 AUGGCCAAGUGGGGAUCUGCUUGCCUCCCAGCU-CACAUACCUGUGG---UGGCGGCGCGUCUUAUUAGAAUUUAUGA-UCCCCAUCU----------------UUCUGCUUUCU ..(((.(((((((((((.(((((((.(((((..-.......))).))---.))))).)).(((....)))......))-)))))).))----------------)...))).... ( -31.60) >DroYak_CAF1 41274 94 + 1 AUGGCCAAGUGGGGAUCUGCUUGCCUCCCAGCU-CACAUACCUGUGG---UGGCGGCGCGUCUUAUUAGAAUUUAUGA-UCCCCAUCU----------------UUUUGCUUUCU ..(((.(((((((((((.(((((((.(((((..-.......))).))---.))))).)).(((....)))......))-)))))).))----------------)...))).... ( -31.60) >DroAna_CAF1 32893 87 + 1 AUGGCACAGUGG-AAUCAGCCU-CGUAUAGGCUAUAUAUACCUGUGG---UGGCGGCGCGUCUUAUUAGAAUUUAUGAGUUGCCAUCU----------------UUUU------- ....(((((((.-....(((((-.....))))).....)).)))))(---(((((((.(((((....))).....)).))))))))..----------------....------- ( -22.70) >consensus AUGGCCAAGUGGGGAUCUGCUUGCCUCCCAGCU_CACAUACCUGUGG___UGGCGGCGCGUCUUAUUAGAAUUUAUGA_UUCCCAUCU________________UUUUGCUUUCU ........(((((((((.(((((((.(((((..........))).))....))))).)).(((....)))......)).)))))))............................. (-17.16 = -17.28 + 0.12)

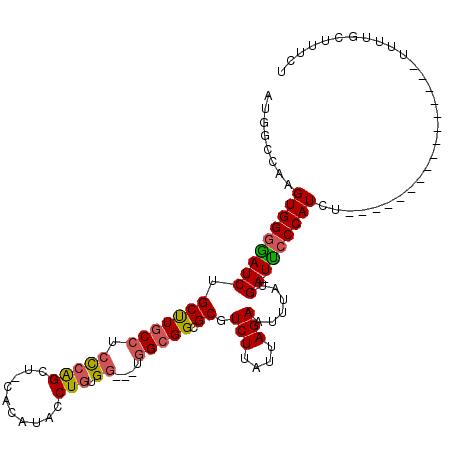

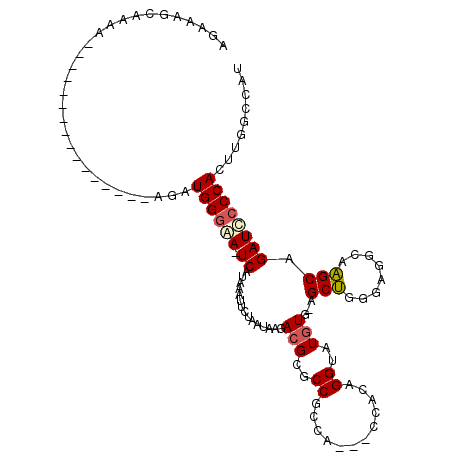
| Location | 14,907,610 – 14,907,714 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.74 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.74 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14907610 104 - 27905053 AGAAAGCAAAAAAGAAAA---------AGAUGGGAA-UCAUAAAUUCUAAUAAGACGCGCCGCCACCACCACAGGUAUGUG-AGCUGGGAGGCAAGCAGAUCCCCACUUGGCCAU .................(---------((.((((.(-((......(((....))).((...(((.((((((((....))))-.).)))..)))..)).))).)))))))...... ( -25.20) >DroSim_CAF1 34896 113 - 1 AGAAAGCAAAAAAAAAAAAGAAAAAAUAGAUGGGAA-UCAUAAAUUCUAAUAAGACGCGCCGCCACCACCACAGGUAUGUG-AGCUGGGAGGCAAGCAGAUCCCCACUUGGCCAU ..........................(((.((((.(-((......(((....))).((...(((.((((((((....))))-.).)))..)))..)).))).)))).)))..... ( -23.80) >DroEre_CAF1 34363 94 - 1 AGAAAGCAGAA----------------AGAUGGGGA-UCAUAAAUUCUAAUAAGACGCGCCGCCA---CCACAGGUAUGUG-AGCUGGGAGGCAAGCAGAUCCCCACUUGGCCAU .....((...(----------------((.((((((-((......(((....))).(((((.(((---(((((....))))-.).)))..)))..)).))))))))))).))... ( -33.10) >DroYak_CAF1 41274 94 - 1 AGAAAGCAAAA----------------AGAUGGGGA-UCAUAAAUUCUAAUAAGACGCGCCGCCA---CCACAGGUAUGUG-AGCUGGGAGGCAAGCAGAUCCCCACUUGGCCAU .....((...(----------------((.((((((-((......(((....))).(((((.(((---(((((....))))-.).)))..)))..)).))))))))))).))... ( -33.10) >DroAna_CAF1 32893 87 - 1 -------AAAA----------------AGAUGGCAACUCAUAAAUUCUAAUAAGACGCGCCGCCA---CCACAGGUAUAUAUAGCCUAUACG-AGGCUGAUU-CCACUGUGCCAU -------....----------------..((((((..........................(((.---.....))).....((((((.....-))))))...-......)))))) ( -17.70) >consensus AGAAAGCAAAA________________AGAUGGGAA_UCAUAAAUUCUAAUAAGACGCGCCGCCA___CCACAGGUAUGUG_AGCUGGGAGGCAAGCAGAUCCCCACUUGGCCAU ..............................((((((.((...............(((..((............))..)))...(((........))).))))))))......... (-14.42 = -14.74 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:39 2006