
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,903,706 – 14,903,801 |
| Length | 95 |
| Max. P | 0.935557 |

| Location | 14,903,706 – 14,903,801 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 89.82 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -23.06 |
| Energy contribution | -23.39 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
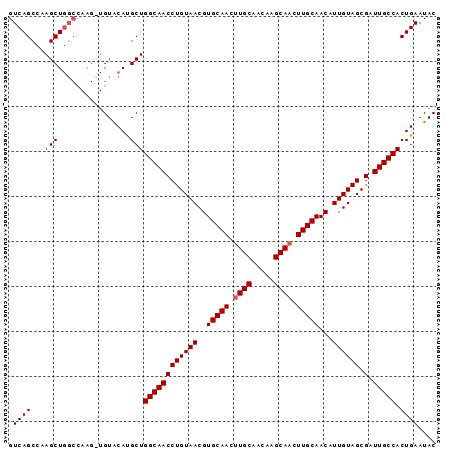
>3R_DroMel_CAF1 14903706 95 + 27905053 GUCAGCCAAGCUGGCCAAGCUGUACACGCUGGCAACCUGUAACGUGCAACGUGCAAUAAGCAACUUGCAACAUUGUAGCGAUUGCCACUGAAUAC ((((((...))))))...........((.(((((((((((((..(((((..(((.....)))..)))))...)))))).).)))))).))..... ( -26.50) >DroSec_CAF1 31123 95 + 1 GUCAGCCAAGCUGGCCAAGUUGUGCAUGCUGGCAACCUGUAACGUGCAACUUGCAACAAGCAACUUGCAACAUUGUAGCGAUUGCCACUGAAUAC ((((((...))))))(((((((..(.(((.(....)..)))..)..)))))))......((((.((((.((...)).)))))))).......... ( -27.30) >DroSim_CAF1 31031 86 + 1 GUCAGCCAAGCU---------GUACAUGCUGGCAACCUGUAACGUGCAACUUGCAACCAGCAACUUGCAACAUUGUAGCGAUUGCCACUGAAUAC .((((...(((.---------......)))((((((((((((..(((((.((((.....)))).)))))...)))))).).))))).)))).... ( -25.00) >consensus GUCAGCCAAGCUGGCCAAG_UGUACAUGCUGGCAACCUGUAACGUGCAACUUGCAACAAGCAACUUGCAACAUUGUAGCGAUUGCCACUGAAUAC .((((...(((................)))((((((((((((..(((((.((((.....)))).)))))...)))))).).))))).)))).... (-23.06 = -23.39 + 0.33)


| Location | 14,903,706 – 14,903,801 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 89.82 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -29.29 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
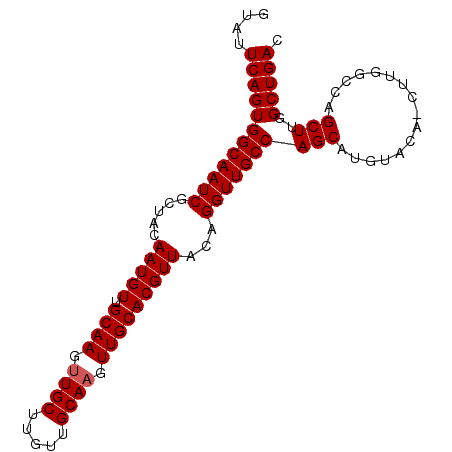
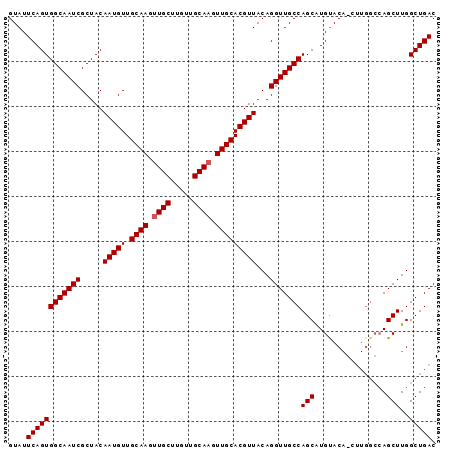
>3R_DroMel_CAF1 14903706 95 - 27905053 GUAUUCAGUGGCAAUCGCUACAAUGUUGCAAGUUGCUUAUUGCACGUUGCACGUUACAGGUUGCCAGCGUGUACAGCUUGGCCAGCUUGGCUGAC ((((.(..((((((((.....(((((.((((..(((.....)))..)))))))))...))))))))..).))))...(..(((.....)))..). ( -32.20) >DroSec_CAF1 31123 95 - 1 GUAUUCAGUGGCAAUCGCUACAAUGUUGCAAGUUGCUUGUUGCAAGUUGCACGUUACAGGUUGCCAGCAUGCACAACUUGGCCAGCUUGGCUGAC ((((.(..((((((((.....(((((.((((.((((.....)))).)))))))))...))))))))).)))).....(..(((.....)))..). ( -34.00) >DroSim_CAF1 31031 86 - 1 GUAUUCAGUGGCAAUCGCUACAAUGUUGCAAGUUGCUGGUUGCAAGUUGCACGUUACAGGUUGCCAGCAUGUAC---------AGCUUGGCUGAC ....((((((((((((.....(((((.((((.((((.....)))).)))))))))...)))))))(((......---------.)))..))))). ( -31.50) >consensus GUAUUCAGUGGCAAUCGCUACAAUGUUGCAAGUUGCUUGUUGCAAGUUGCACGUUACAGGUUGCCAGCAUGUACA_CUUGGCCAGCUUGGCUGAC ....((((((((((((.....(((((.((((.((((.....)))).)))))))))...)))))))(((................)))..))))). (-29.29 = -29.62 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:35 2006