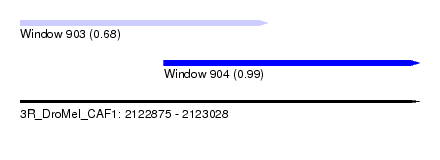
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,122,875 – 2,123,028 |
| Length | 153 |
| Max. P | 0.992177 |

| Location | 2,122,875 – 2,122,970 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.24 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.65 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
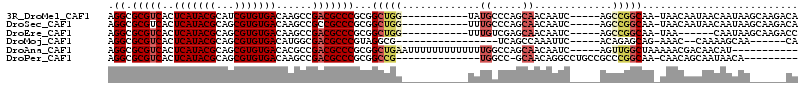
>3R_DroMel_CAF1 2122875 95 + 27905053 UUUAUUUUGGCC--------------GCUUUUGGCCGUUUUACGCGGCUAAAACAAUAACCGAAAAUAAAGGCGCGUCACUCAUACGCAUCGUGUGACAAGCCGACGCC ((((((((((..--------------..((((((((((.....))))))))))......)).))))))))((((((.(..(((((((...)))))))...).)).)))) ( -30.20) >DroSec_CAF1 265959 93 + 1 --UUUUUGGGCC--------------ACUUUUGGCCGUUUUACGCGGCUAAAACAAUAACCGAAAAUAAAGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGCCGCC --(((((((...--------------..((((((((((.....))))))))))......)))))))....((((.((...(((((((...)))))))...))))))... ( -33.40) >DroSim_CAF1 284582 95 + 1 UUUUUUUGGGCC--------------GCUUUUGGCCGUUUUACGCGGCUAAAACAAUAACCGAAAAUAAAGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGCCGCC ..(((((((...--------------..((((((((((.....))))))))))......)))))))....((((.((...(((((((...)))))))...))))))... ( -33.60) >DroEre_CAF1 268973 92 + 1 U---CUCGGACU--------------GCUUUUGGCCGUUUUACGCGGCUAAAACAAUAACCGAAAAUAAAGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGACGCC .---.((((..(--------------(.((((((((((.....))))))))))))....)))).......((((((.(..(((((((...)))))))...).)).)))) ( -30.90) >DroWil_CAF1 267483 109 + 1 UUCUUUCCCUCCGCAACUCCUCCCUUGCUUUGCUUUGUUUUACGCAGCUAAAACAAUAACCGAAAAUAAAGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGACGCC ...((((.....((((........))))......((((((((......)))))))).....)))).....((((((.(..(((((((...)))))))...).)).)))) ( -21.60) >DroYak_CAF1 270153 91 + 1 U----UCGGGCC--------------GCUUUUGGCCGUUUUACGCGGCUAAAACAAUAACCGAAAAUAAAGGCGUGUCACUCAUACGCAGCGUGUGACAAGCCGACGCC (----((((...--------------..((((((((((.....))))))))))......)))))......(((.((((((.(.........).)))))).)))...... ( -32.40) >consensus U_UUUUCGGGCC______________GCUUUUGGCCGUUUUACGCGGCUAAAACAAUAACCGAAAAUAAAGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGACGCC ............................((((((((((.....)))))))))).................((((((.(..(((((((...)))))))...).)).)))) (-23.70 = -23.65 + -0.05)


| Location | 2,122,930 – 2,123,028 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.96 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -16.94 |
| Energy contribution | -17.89 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
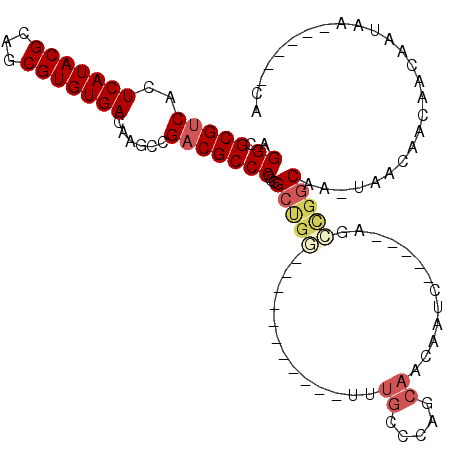
>3R_DroMel_CAF1 2122930 98 + 27905053 AGGCGCGUCACUCAUACGCAUCGUGUGACAAGCCGACGCCCGCGGCUGG-----------UAUGCCCAGCAACAAUC-----AGCCGGCAA-UAACAAUAACAAUAAGCAAGACA .((.(((((..(((((((...)))))))......)))))))((((((((-----------(.(((...)))...)))-----)))).))..-....................... ( -29.40) >DroSec_CAF1 266012 98 + 1 AGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGCCGCCCGCGGCUGG-----------UUUGCCCAGCAACAAUC-----AGCCGGCAA-UAACAAUAACAAUAAGCAAGACA .((((.((...(((((((...)))))))...))))))(((...((((((-----------(((((...)))..))))-----)))))))..-....................... ( -31.60) >DroEre_CAF1 269025 92 + 1 AGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGACGCCCGCGGCUGG-----------UUUGUCGAGCAACAAUC-----AGCCGGCAA-UAA------CAAUAAGCAAGACC .((.(((((..(((((((...)))))))......)))))))((((((((-----------((.((......))))))-----)))).))..-...------.............. ( -29.90) >DroMoj_CAF1 279818 84 + 1 AGGCGCGUCACUCAUACGCAGCGUGUGACAUGGCGACGCCCGUAGGCG-----------------UCAGCCAAAUUC-----ACAGAGCAG-AAAC--CAAAAGCAA------CA ..((((((.......)))).((.(((((..((((((((((....))))-----------------)).))))...))-----)))..))..-....--.....))..------.. ( -31.80) >DroAna_CAF1 238900 99 + 1 AGGCGCGUCACUCAUACGCAGCGUGUGACACGCCGACGCCCGCGGCUGAAUUUUUUUUUUUUUGGCCAGCAACAAUC-----AGUUGGCUAAAAACGACAACAU----------- .((((.(((((.(.........).))))).))))...(((...)))............((((((((((((.......-----.)))))))))))).........----------- ( -32.90) >DroPer_CAF1 241113 90 + 1 AGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGACGCCCGCGGCCG--------------UGGCC-GCAACAGGCCUGCCGCCCGGCAA-CAACAGCAAUAACA--------- .(((..(((((.(.........).)))))..)))...(((.(((((..--------------.((((-......)))).)))))..)))..-..............--------- ( -35.80) >consensus AGGCGCGUCACUCAUACGCAGCGUGUGACAAGCCGACGCCCGCGGCUGG___________UUUGCCCAGCAACAAUC_____AGCCGGCAA_UAACAACAACAAUAA______CA .((.(((((..(((((((...)))))))......)))))))...(((((.............((.....)).............))))).......................... (-16.94 = -17.89 + 0.95)
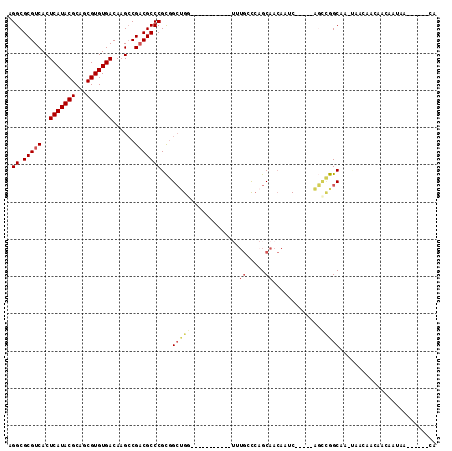
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:17 2006