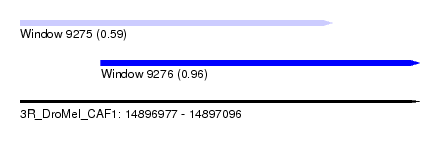
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,896,977 – 14,897,096 |
| Length | 119 |
| Max. P | 0.956612 |

| Location | 14,896,977 – 14,897,070 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.83 |
| Mean single sequence MFE | -19.37 |
| Consensus MFE | -13.59 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
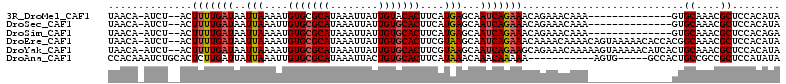
>3R_DroMel_CAF1 14896977 93 + 27905053 -----------GAUGGCAAUGAACUU-----AUAUUCGUUUAACA-AUCU--ACUUUUGAUAAUUAAAAUGUGCGCAUAAAUUAUUGUACACUUCAUGAGCAAUCAGAAACA -----------(((.((.(((((...-----.......(((((..-(((.--......)))..)))))..(((..((........))..))))))))..)).)))....... ( -14.30) >DroSec_CAF1 24565 93 + 1 -----------GAUGGCAAUGAACUU-----AACUUCGUUUAACA-AUCU--ACUUUUGAUAAUUAAAUUGUGCGCAUAAAUUAUUGUGCACUUCAUGAGCAAUCAGAAACA -----------(((.((.(((((...-----......((((((..-(((.--......)))..)))))).(((((((........))))))))))))..)).)))....... ( -20.50) >DroSim_CAF1 24435 98 + 1 -----------GAUGGCAAUGAACUUAACUUAACUUCGUUUAACA-AUCU--ACUUUUGAUAAUUAAAAUGUGCGCAUAAAUUAUUGUGCACUUCAUGAGCAAUCAGAAACA -----------(((.((.(((((...............(((((..-(((.--......)))..)))))..(((((((........))))))))))))..)).)))....... ( -19.70) >DroEre_CAF1 24064 93 + 1 -----------GGUGGCAAUGAACUU-----AACUUCAUUUAACA-AUCU--ACUUUUGAUAAUUAAAAUGUGCGCAUAAAUUAUUGUGCACUUCGUAAGCAAUCAGAAACA -----------.((...((((((...-----...))))))..)).-....--..(((((((..(((....(((((((........)))))))....)))...)))))))... ( -19.90) >DroYak_CAF1 30602 93 + 1 -----------GGUGGCAAUGAACUU-----AACUUCAUUUAACA-AUCU--ACUUUUGAUAAUUAAAAUGUGCGCAUAAAUUAUUGUGCACUUCGUAAGCAAUCAGAAGCA -----------.((...((((((...-----...))))))..)).-....--.((((((((..(((....(((((((........)))))))....)))...)))))))).. ( -22.50) >DroAna_CAF1 23371 105 + 1 AGUCACUGCCGAAUGGCAUUGAACCU-----AACGUAGCCCCACAAAUCUGCACUCUUGAUUAUUAAAUUGUGCGCAUAAAUUACUGUGCACUUCAUAAACAAACAAAAA-- ..(((.((((....)))).)))....-----...((((..........))))....(((.((((......(((((((........)))))))...)))).))).......-- ( -19.30) >consensus ___________GAUGGCAAUGAACUU_____AACUUCGUUUAACA_AUCU__ACUUUUGAUAAUUAAAAUGUGCGCAUAAAUUAUUGUGCACUUCAUAAGCAAUCAGAAACA ......................................................(((((((..(((....(((((((........)))))))....)))...)))))))... (-13.59 = -13.70 + 0.11)



| Location | 14,897,001 – 14,897,096 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -18.42 |
| Consensus MFE | -13.99 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14897001 95 + 27905053 UAACA-AUCU--ACUUUUGAUAAUUAAAAUGUGCGCAUAAAUUAUUGUACACUUCAUGAGCAAUCAGAAACAGAAACAAA--------------GUGCAAACGCUCCACAUA .....-.(((--..(((((((..(((....(((..((........))..)))....)))...)))))))..))).....(--------------(((....))))....... ( -13.50) >DroSec_CAF1 24589 95 + 1 UAACA-AUCU--ACUUUUGAUAAUUAAAUUGUGCGCAUAAAUUAUUGUGCACUUCAUGAGCAAUCAGAAACAGAAACAAA--------------GUGCAAACGCUCCACAUA .....-.(((--..(((((((..(((....(((((((........)))))))....)))...)))))))..))).....(--------------(((....))))....... ( -18.90) >DroSim_CAF1 24464 95 + 1 UAACA-AUCU--ACUUUUGAUAAUUAAAAUGUGCGCAUAAAUUAUUGUGCACUUCAUGAGCAAUCAGAAACAGAAACAAA--------------GUGCAAACGCUCCACAGA .....-.(((--..(((((((..(((....(((((((........)))))))....)))...)))))))..))).....(--------------(((....))))....... ( -18.90) >DroEre_CAF1 24088 109 + 1 UAACA-AUCU--ACUUUUGAUAAUUAAAAUGUGCGCAUAAAUUAUUGUGCACUUCGUAAGCAAUCAGAAACAAAACAAAACAGUAAAAACACCACGGCAAACGCUCCACAUA .....-....--..(((((((..(((....(((((((........)))))))....)))...)))))))..........................(((....)))....... ( -17.70) >DroYak_CAF1 30626 109 + 1 UAACA-AUCU--ACUUUUGAUAAUUAAAAUGUGCGCAUAAAUUAUUGUGCACUUCGUAAGCAAUCAGAAGCAGAAACAAAAAGUAAAAACAUCACUGCAAACGCUCCACAUA .....-.(((--.((((((((..(((....(((((((........)))))))....)))...)))))))).)))......................((....))........ ( -22.60) >DroAna_CAF1 23406 96 + 1 CCACAAAUCUGCACUCUUGAUUAUUAAAUUGUGCGCAUAAAUUACUGUGCACUUCAUAAACAAACAAAAA-----------AGUG-----GCCACUGCCGCCGCUCCAUAUA ..........((....(((.((((......(((((((........)))))))...)))).))).......-----------.(((-----((....))))).))........ ( -18.90) >consensus UAACA_AUCU__ACUUUUGAUAAUUAAAAUGUGCGCAUAAAUUAUUGUGCACUUCAUAAGCAAUCAGAAACAGAAACAAA______________CUGCAAACGCUCCACAUA ..............(((((((..(((....(((((((........)))))))....)))...)))))))...........................((....))........ (-13.99 = -14.10 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:25 2006