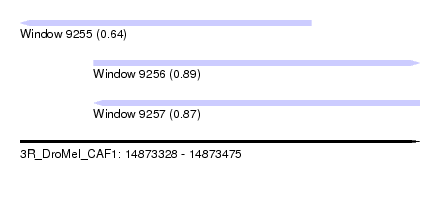
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,873,328 – 14,873,475 |
| Length | 147 |
| Max. P | 0.886406 |

| Location | 14,873,328 – 14,873,435 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.44 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14873328 107 - 27905053 CACGGUAUAAAUAUCGAGCACCUAACGCACAAGCAACGUAAGUGACUUGUUUAGCUAAAGCAUAAAAAUCACAAUCAUAAGUCACUUUUUUC-------------UUGAUCGUGCUAAUU ..(((((....)))))((((((....)..((((.((...((((((((((((((((....)).)))...........)))))))))))..)))-------------)))...))))).... ( -22.80) >DroSec_CAF1 991 107 - 1 CACGGUAUAAAUAUCGAGAACCUAACGCACAAGCAACGUAAGUGACUUGUUUAGCUAAAGCAUAAAACUCACAAUCAUAAGUCACUUUUUUC-------------UUGAUCGUGCUAUUU ...(((((....((((((((......((....)).....((((((((((((((((....)).)))...........)))))))))))..)))-------------))))).))))).... ( -22.40) >DroSim_CAF1 993 107 - 1 CACGGUAUAAAUAUCGAGCACCUAACGCACAAGCAACGUAAGUGACUUAUUCAGCUAAAGCAUAAAAAUCACAAUCAUAAGUCACUUUUUUC-------------UUGAUCGUGCUAUUU ..(((((....)))))((((((....)..((((.((...(((((((((((...((....))...............)))))))))))..)))-------------)))...))))).... ( -21.47) >DroEre_CAF1 987 119 - 1 CACGGUAUAAAAAUCGAGCUCCUAACGCACAAGCAACGUAAGUGACUUGUUUAGCUAAAGCAUACAAAUCACAAUCAUAAGUCACUUUUUUCAA-UUGUUAAAAGAUUGUCGUGCUCUUU ..((((......))))..........((((..((((.....((((.((((...((....))..)))).))))............(((((..(..-..)..))))).)))).))))..... ( -20.90) >DroYak_CAF1 988 120 - 1 CACGGUAUAAAUAUCGAGCCCCAAGCGCACACGAAAAGUAAGUGACUUGUUUAGCUAAAGCAUACAAAUCACAAUCAUAAGUCACUUUUUUCAAAAUAUUACUAGUUUGUCGUGCUAUUU ..(((((....))))).((.....))(((((((((.(((((((((.((((...((....))..)))).))))......(((...)))...........)))))..))))).))))..... ( -22.70) >consensus CACGGUAUAAAUAUCGAGCACCUAACGCACAAGCAACGUAAGUGACUUGUUUAGCUAAAGCAUAAAAAUCACAAUCAUAAGUCACUUUUUUC_____________UUGAUCGUGCUAUUU ..(((((....)))))..........((((...(((...(((((((((((...((....))...............)))))))))))..................)))...))))..... (-15.65 = -15.85 + 0.20)

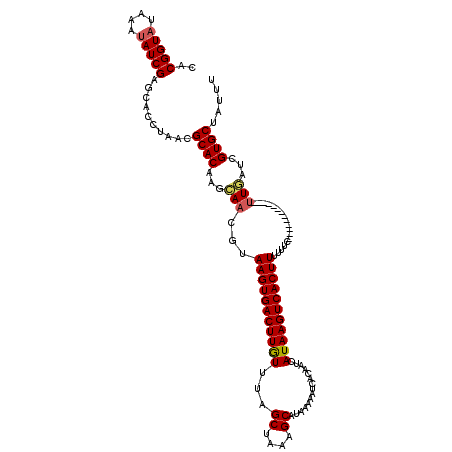

| Location | 14,873,355 – 14,873,475 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.52 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14873355 120 + 27905053 UUAUGAUUGUGAUUUUUAUGCUUUAGCUAAACAAGUCACUUACGUUGCUUGUGCGUUAGGUGCUCGAUAUUUAUACCGUGGCUGUGUUUAACCUGGGGUAUGCCGAAAUAAUUAUCAUAU .((((((....(((((((((((((((.((((((((((((..((((.......))))..((((...........)))))))))).))))))..))))))))))..)))))....)))))). ( -31.10) >DroSec_CAF1 1018 120 + 1 UUAUGAUUGUGAGUUUUAUGCUUUAGCUAAACAAGUCACUUACGUUGCUUGUGCGUUAGGUUCUCGAUAUUUAUACCGUGGCUGUGUUUAACCUGGGGUAUGCCGAAAUAAUUAUCAUAU .((((((.((...(((((((((((((.((((((((((((..((((.......))))..(((.............))))))))).))))))..))))))))))..)))...)).)))))). ( -29.02) >DroSim_CAF1 1020 120 + 1 UUAUGAUUGUGAUUUUUAUGCUUUAGCUGAAUAAGUCACUUACGUUGCUUGUGCGUUAGGUGCUCGAUAUUUAUACCGUGGCUGUGUUUAACCUGGGGUAUGCCGAAAUAAUUAUCAUAU .((((((....(((((((((((((((.((((((((((((..((((.......))))..((((...........)))))))))).))))))..))))))))))..)))))....)))))). ( -29.30) >DroEre_CAF1 1026 120 + 1 UUAUGAUUGUGAUUUGUAUGCUUUAGCUAAACAAGUCACUUACGUUGCUUGUGCGUUAGGAGCUCGAUUUUUAUACCGUGGCUGUGUUUAACCUGGGGUAUGCCGAAAUAAUUAUCAUAU .((((((....(((((((((((((((.((((((((((((...((..((((.((...)).)))).))((....))...)))))).))))))..)))))))))))..))))....)))))). ( -30.80) >DroYak_CAF1 1028 120 + 1 UUAUGAUUGUGAUUUGUAUGCUUUAGCUAAACAAGUCACUUACUUUUCGUGUGCGCUUGGGGCUCGAUAUUUAUACCGUGGCUGUGUUUAACCUGGGGUAUGCCGAAAUAAGUAUCAUAU .(((((..(((((((((..((....))...))))))))).((((((((((((((.((..((....(((((.............)))))...))..))))))).))))..)))))))))). ( -32.52) >consensus UUAUGAUUGUGAUUUUUAUGCUUUAGCUAAACAAGUCACUUACGUUGCUUGUGCGUUAGGUGCUCGAUAUUUAUACCGUGGCUGUGUUUAACCUGGGGUAUGCCGAAAUAAUUAUCAUAU .((((((....(((((((((((((((.((((((((((((......(((....)))...((...............)))))))).))))))..))))))))))..)))))....)))))). (-26.04 = -26.52 + 0.48)


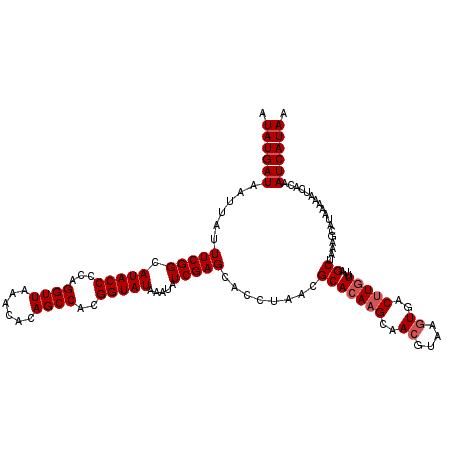
| Location | 14,873,355 – 14,873,475 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -23.54 |
| Consensus MFE | -18.26 |
| Energy contribution | -18.86 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14873355 120 - 27905053 AUAUGAUAAUUAUUUCGGCAUACCCCAGGUUAAACACAGCCACGGUAUAAAUAUCGAGCACCUAACGCACAAGCAACGUAAGUGACUUGUUUAGCUAAAGCAUAAAAAUCACAAUCAUAA .((((((....((((..(((((((...((((......))))..)))))........(((..(....).(((((..((....))..)))))...)))...))....))))....)))))). ( -22.40) >DroSec_CAF1 1018 120 - 1 AUAUGAUAAUUAUUUCGGCAUACCCCAGGUUAAACACAGCCACGGUAUAAAUAUCGAGAACCUAACGCACAAGCAACGUAAGUGACUUGUUUAGCUAAAGCAUAAAACUCACAAUCAUAA .((((((............(((((...((((......))))..))))).......(((..........(((((..((....))..)))))...((....))......)))...)))))). ( -24.00) >DroSim_CAF1 1020 120 - 1 AUAUGAUAAUUAUUUCGGCAUACCCCAGGUUAAACACAGCCACGGUAUAAAUAUCGAGCACCUAACGCACAAGCAACGUAAGUGACUUAUUCAGCUAAAGCAUAAAAAUCACAAUCAUAA .((((((..((((((((((........((((......)))).(((((....))))).)).......((....))..)).))))))........((....))............)))))). ( -19.40) >DroEre_CAF1 1026 120 - 1 AUAUGAUAAUUAUUUCGGCAUACCCCAGGUUAAACACAGCCACGGUAUAAAAAUCGAGCUCCUAACGCACAAGCAACGUAAGUGACUUGUUUAGCUAAAGCAUACAAAUCACAAUCAUAA .((((((..((((....(((((((...((((......))))..))))).........((.......))....))...))))((((.((((...((....))..)))).)))).)))))). ( -24.60) >DroYak_CAF1 1028 120 - 1 AUAUGAUACUUAUUUCGGCAUACCCCAGGUUAAACACAGCCACGGUAUAAAUAUCGAGCCCCAAGCGCACACGAAAAGUAAGUGACUUGUUUAGCUAAAGCAUACAAAUCACAAUCAUAA .((((((((((..(((((((((((...((((......))))..))))).........((.....))))...))))))))).((((.((((...((....))..)))).))))..))))). ( -27.30) >consensus AUAUGAUAAUUAUUUCGGCAUACCCCAGGUUAAACACAGCCACGGUAUAAAUAUCGAGCACCUAACGCACAAGCAACGUAAGUGACUUGUUUAGCUAAAGCAUAAAAAUCACAAUCAUAA .((((((......(((((.(((((...((((......))))..))))).....)))))........(((((((..((....))..)))))...))..................)))))). (-18.26 = -18.86 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:07 2006