
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,849,969 – 14,850,249 |
| Length | 280 |
| Max. P | 0.999868 |

| Location | 14,849,969 – 14,850,089 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -38.85 |
| Consensus MFE | -34.12 |
| Energy contribution | -33.80 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14849969 120 - 27905053 ACCUUUAUGUUCUUGGCAGUAUCUUCGGCCACUUUUUGCGCCAGUUUUCCGCUGUCGAGAGCAGUUUGCAACUGCUGCACACACCAGCAGAGCUCCACUUCGAACUGGGUGACAAGGUCG (((((..(((((((((((((.....((((((.....)).))).)......)))))))))))))...((((.....))))(((.((((..(((......)))...)))))))..))))).. ( -37.30) >DroSec_CAF1 3206 120 - 1 ACCUUUAUGUUCUUGGCGGUAUCUUCGGCCACUUUCUGGGCCAAUUUUCCGCUGUCGAGAGCAGUUUGCAACUGCUGCACACACCAGCAGAGUUCCACUUCGAACUGGGUGACGAGAUCG .......(((((((((((((......((((.(.....)))))........)))))))))))))...((.((((.((((........)))))))).))...(((.((.(....).)).))) ( -39.54) >DroSim_CAF1 4018 120 - 1 ACCUUUAUGUUCUUGGCGGUAUCUUCGGCCACUUUCUGGGCCAGUUUUCCGCUGUCGAGAGCAGUUUGCAACUGCUGCACACACCAGCAGAGUUCCACUUCGAGCUGGGUGACGAGAUCG .......(((((((((((((.....(((((.(.....))))).)......)))))))))))))...((((.....))))(((.(((((.(((......)))..))))))))......... ( -40.30) >DroEre_CAF1 3377 120 - 1 ACCUUUAUGUUCUUGGCGGUAUCUUCGGCCACUUUCUGGGCCAGUUUUCCGCUUUCAAGGGCGGUUUGCAACUGCUGCACACACCAGCAGAGCUCCACUUCAAACUGGGUGACGAGGUCC ..........(((((((((((.....((((.(.....))))).((...((((((....))))))...))...)))))).(((.((((..(((......)))...))))))).)))))... ( -37.60) >DroYak_CAF1 3814 120 - 1 ACCUUUAUGUUCUUGGCGGUAUCUUCGGCCACUUUCUGGGCCAGCUUUCCGCUUUCAAGAGCGGUUUUCAACUGCUGCACACACCAGCAGAGCUCCACUUCGAACUGGGUGACAAGGUUU (((((...((((..((.((.......((((.(.....)))))((((..((((((....)))))).......((((((.......)))))))))))).))..))))..(....)))))).. ( -39.50) >consensus ACCUUUAUGUUCUUGGCGGUAUCUUCGGCCACUUUCUGGGCCAGUUUUCCGCUGUCGAGAGCAGUUUGCAACUGCUGCACACACCAGCAGAGCUCCACUUCGAACUGGGUGACGAGGUCG ((((....((((((((((((......((((........))))........))))))))))))((((((...((((((.......))))))((.....)).)))))))))).......... (-34.12 = -33.80 + -0.32)



| Location | 14,850,009 – 14,850,129 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -31.48 |
| Energy contribution | -31.92 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14850009 120 + 27905053 GUGCAGCAGUUGCAAACUGCUCUCGACAGCGGAAAACUGGCGCAAAAAGUGGCCGAAGAUACUGCCAAGAACAUAAAGGUACUAACCAGUUCAACAGCGCCGCUCAUCAGGAAGCGGCAG ..((.(((((..(...(((((......))))).....((((.((.....))))))..)..)))))...((((.....(((....))).))))....))((((((........)))))).. ( -33.90) >DroSec_CAF1 3246 120 + 1 GUGCAGCAGUUGCAAACUGCUCUCGACAGCGGAAAAUUGGCCCAGAAAGUGGCCGAAGAUACCGCCAAGAACAUAAAGGUGCUGACCAGCUCAACAGCGCCGCUCAUCAGGAAGCGGCAG (.((((((((.....)))))).....((((((....(((((((.....).)))))).....))(((...........)))))))....)).)......((((((........)))))).. ( -37.70) >DroSim_CAF1 4058 120 + 1 GUGCAGCAGUUGCAAACUGCUCUCGACAGCGGAAAACUGGCCCAGAAAGUGGCCGAAGAUACCGCCAAGAACAUAAAGGUGCUGACCAGCUCAACAGCGCCGCUCGUCAGAAAGCGGCAG .(((.(((((.....)))))(((.(((.((((.....((((((.....).)))))......))))............(((((((..........)))))))....)))))).....))). ( -38.60) >DroEre_CAF1 3417 120 + 1 GUGCAGCAGUUGCAAACCGCCCUUGAAAGCGGAAAACUGGCCCAGAAAGUGGCCGAAGAUACCGCCAAGAACAUAAAGGUCCUGACCAGCUCAACGGCGCCGCUCAUCAGGAAGCGGCAG .(((((...)))))....(((.((((..((((.....((((((.....).)))))......))))............(((....)))...)))).)))((((((........)))))).. ( -38.60) >DroYak_CAF1 3854 120 + 1 GUGCAGCAGUUGAAAACCGCUCUUGAAAGCGGAAAGCUGGCCCAGAAAGUGGCCGAAGAUACCGCCAAGAACAUAAAGGUCCUGACCAGCUCAACAGCGCCGCUCAUCAGAAAGCGGCAG .....((.(((((...(((((......)))))...(((((..(((....((((.(......).)))).((.(.....).))))).)))))))))).))((((((........)))))).. ( -40.80) >consensus GUGCAGCAGUUGCAAACUGCUCUCGACAGCGGAAAACUGGCCCAGAAAGUGGCCGAAGAUACCGCCAAGAACAUAAAGGUCCUGACCAGCUCAACAGCGCCGCUCAUCAGGAAGCGGCAG ..((.(((((.....)))))....((..((((.....(((((........)))))......))))............(((....)))...))....))((((((........)))))).. (-31.48 = -31.92 + 0.44)


| Location | 14,850,009 – 14,850,129 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -46.59 |
| Consensus MFE | -42.94 |
| Energy contribution | -42.58 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14850009 120 - 27905053 CUGCCGCUUCCUGAUGAGCGGCGCUGUUGAACUGGUUAGUACCUUUAUGUUCUUGGCAGUAUCUUCGGCCACUUUUUGCGCCAGUUUUCCGCUGUCGAGAGCAGUUUGCAACUGCUGCAC ..(((((((......)))))))((.((((.((((((....))).....((((((((((((.....((((((.....)).))).)......)))))))))))))))...)))).))..... ( -42.80) >DroSec_CAF1 3246 120 - 1 CUGCCGCUUCCUGAUGAGCGGCGCUGUUGAGCUGGUCAGCACCUUUAUGUUCUUGGCGGUAUCUUCGGCCACUUUCUGGGCCAAUUUUCCGCUGUCGAGAGCAGUUUGCAACUGCUGCAC ..(((((((......)))))))((.((((.(((....))).......(((((((((((((......((((.(.....)))))........))))))))))))).....)))).))..... ( -45.94) >DroSim_CAF1 4058 120 - 1 CUGCCGCUUUCUGACGAGCGGCGCUGUUGAGCUGGUCAGCACCUUUAUGUUCUUGGCGGUAUCUUCGGCCACUUUCUGGGCCAGUUUUCCGCUGUCGAGAGCAGUUUGCAACUGCUGCAC ..(((((((......)))))))((.((((.(((....))).......(((((((((((((.....(((((.(.....))))).)......))))))))))))).....)))).))..... ( -46.10) >DroEre_CAF1 3417 120 - 1 CUGCCGCUUCCUGAUGAGCGGCGCCGUUGAGCUGGUCAGGACCUUUAUGUUCUUGGCGGUAUCUUCGGCCACUUUCUGGGCCAGUUUUCCGCUUUCAAGGGCGGUUUGCAACUGCUGCAC ..(((((((......)))))))((.(((((((((((((((((......)))))((((.(......).)))).......))))))))..((((((....))))))....)))).))..... ( -46.60) >DroYak_CAF1 3854 120 - 1 CUGCCGCUUUCUGAUGAGCGGCGCUGUUGAGCUGGUCAGGACCUUUAUGUUCUUGGCGGUAUCUUCGGCCACUUUCUGGGCCAGCUUUCCGCUUUCAAGAGCGGUUUUCAACUGCUGCAC ..(((((((......)))))))((.(((((((((((((((((......)))))((((.(......).)))).......)))))))...((((((....))))))...))))).))..... ( -51.50) >consensus CUGCCGCUUCCUGAUGAGCGGCGCUGUUGAGCUGGUCAGCACCUUUAUGUUCUUGGCGGUAUCUUCGGCCACUUUCUGGGCCAGUUUUCCGCUGUCGAGAGCAGUUUGCAACUGCUGCAC ..(((((((......)))))))((.((((.((((((....))).....((((((((((((......((((........))))........)))))))))))))))...)))).))..... (-42.94 = -42.58 + -0.36)


| Location | 14,850,089 – 14,850,209 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -35.38 |
| Energy contribution | -35.42 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14850089 120 + 27905053 ACUAACCAGUUCAACAGCGCCGCUCAUCAGGAAGCGGCAGGUGAUGAAGCUAGCCCUGGGCGACUACCGAGCGAAGAUGCAGCAGGAGGAGCAGAAGUUGCUACUAGCCUCCAGGCAAAU .......((((((.((.(((((((........)))))).).)).)).)))).(((.((((.(.(((...(((((...(((..........)))....)))))..)))).))))))).... ( -36.10) >DroSec_CAF1 3326 120 + 1 GCUGACCAGCUCAACAGCGCCGCUCAUCAGGAAGCGGCAGGUGAUGAAGCUAGCCCUGGGCGACUACCGAGCGAAGAUGCAGCAGGAGGAGCAGAAAUUGCUACUAGCCUCCAAGCAAAU ((((....((((......((((((........)))))).((((.....(((.......)))...)))))))).......)))).((((((((((...))))).....)))))........ ( -40.10) >DroSim_CAF1 4138 120 + 1 GCUGACCAGCUCAACAGCGCCGCUCGUCAGAAAGCGGCAGGUGAUGAAGCUAGCUCUGGGCGACUACCGAGCGAAGAUGCAGCAGGAGGAGCAGAAGUUGCUACUAGCCUCCAAACAAAU ((((...((((((.((.(((((((........)))))).).)).)).)))).((((.((.......)))))).......)))).((((((((((...))))).....)))))........ ( -43.30) >DroEre_CAF1 3497 120 + 1 CCUGACCAGCUCAACGGCGCCGCUCAUCAGGAAGCGGCAGGUGAUGAAGCUAGCCCUGGGCGACUACCGAGCGAAGAUGCAGCAGGAGGAGCAGAAAAUGCUUCUAGCCUCCCGGCAAAU ((((....(((((..(((((((((........)))))).(((......))).))).))))).........((......))..))))(((((((.....))))))).(((....))).... ( -43.00) >DroYak_CAF1 3934 120 + 1 CCUGACCAGCUCAACAGCGCCGCUCAUCAGAAAGCGGCAGGUGAUGAAGCUAGCCCUUGGCGACUACCGAGCGAAGAUGCAGCAGGAGGAGCAGAAAAUGCUUCUAGCCUCCAGGCAAAU ((((...((((((.((.(((((((........)))))).).)).)).)))).((.(((.((.........)).)))..))..))))(((((((.....))))))).(((....))).... ( -43.50) >consensus CCUGACCAGCUCAACAGCGCCGCUCAUCAGGAAGCGGCAGGUGAUGAAGCUAGCCCUGGGCGACUACCGAGCGAAGAUGCAGCAGGAGGAGCAGAAAUUGCUACUAGCCUCCAGGCAAAU ........((((......((((((........)))))).((((.....(((.......)))...)))))))).....(((....(((((((((.....)))).....)))))..)))... (-35.38 = -35.42 + 0.04)

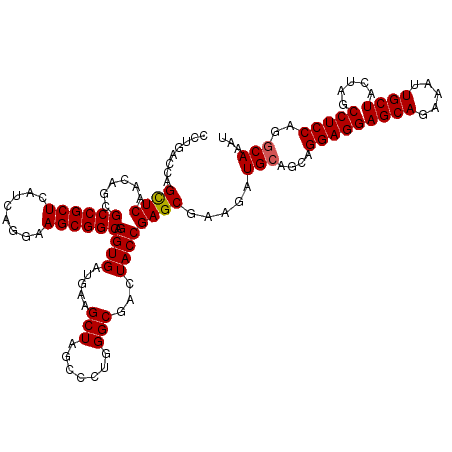

| Location | 14,850,129 – 14,850,249 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -32.76 |
| Energy contribution | -32.88 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14850129 120 - 27905053 GCUAGACUUUUUGGAUGAUUCAGCCGUAGGUGUGAAUUUGAUUUGCCUGGAGGCUAGUAGCAACUUCUGCUCCUCCUGCUGCAUCUUCGCUCGGUAGUCGCCCAGGGCUAGCUUCAUCAC (((((.((..((((.(((((..((((.....(((((..((....((..(((((..(((((......)))))))))).))..))..))))).))))))))).)))))))))))........ ( -39.90) >DroSec_CAF1 3366 120 - 1 GCUAGACUUUUUGGAUGAUUCAGCCGUUGGUGUGAAUUUGAUUUGCUUGGAGGCUAGUAGCAAUUUCUGCUCCUCCUGCUGCAUCUUCGCUCGGUAGUCGCCCAGGGCUAGCUUCAUCAC (((((.((..((((.(((((..((((.....(((((..((....((..(((((..(((((......)))))))))).))..))..))))).))))))))).)))))))))))........ ( -40.00) >DroSim_CAF1 4178 120 - 1 GCUAGACUUUUUGGAUGAUUCAGCCGUUGGUGUGAAUUUGAUUUGUUUGGAGGCUAGUAGCAACUUCUGCUCCUCCUGCUGCAUCUUCGCUCGGUAGUCGCCCAGAGCUAGCUUCAUCAC (((((.(((..(((.(((((..((((.....(((((..((....((..(((((..(((((......)))))))))).))..))..))))).))))))))).)))))))))))........ ( -39.00) >DroEre_CAF1 3537 120 - 1 GCUAGACUUUUUGGAUGCUUCAGCCGUAGGUGUGAAUUUGAUUUGCCGGGAGGCUAGAAGCAUUUUCUGCUCCUCCUGCUGCAUCUUCGCUCGGUAGUCGCCCAGGGCUAGCUUCAUCAC (((((.((..((((..((..(.((((.....(((((..((....((.((((((.((((((...))))))..))))))))..))..))))).)))).)..)))))))))))))........ ( -37.40) >DroYak_CAF1 3974 120 - 1 GCUAGACUUUUUGGAUGCUUCAGUCGUAGGUGUAAAUUUGAUUUGCCUGGAGGCUAGAAGCAUUUUCUGCUCCUCCUGCUGCAUCUUCGCUCGGUAGUCGCCAAGGGCUAGCUUCAUCAC (((((.(((((.((..(((((((.((.(((((((..........((..(((((.((((((...))))))..))))).))))))))).)))).)).)))..))))))))))))........ ( -35.10) >consensus GCUAGACUUUUUGGAUGAUUCAGCCGUAGGUGUGAAUUUGAUUUGCCUGGAGGCUAGUAGCAAUUUCUGCUCCUCCUGCUGCAUCUUCGCUCGGUAGUCGCCCAGGGCUAGCUUCAUCAC (((((.((((..((.(((((..((((.....(((((..((....((..(((((.....((((.....))))))))).))..))..))))).))))))))))).)))))))))........ (-32.76 = -32.88 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:59 2006