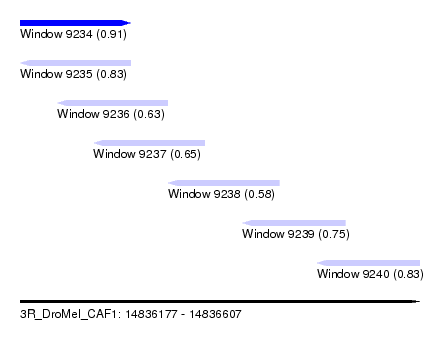
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,836,177 – 14,836,607 |
| Length | 430 |
| Max. P | 0.908321 |

| Location | 14,836,177 – 14,836,296 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -35.52 |
| Energy contribution | -35.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14836177 119 + 27905053 AGCCACUAUCUCAAAGAUGGCCCAUUUGGGCGGCUCUCGCUGUCGUUGUUGUUGCCGUAUUUGC-CGUCUUGACGGCACUUAAAUUGCAUUAUUAUAUACGGCUCGACAUUUAUGUGCAA ((((..((((.....))))((((....))))))))...((......(((((..(((((((.(((-(((....))))))....(((......)))..))))))).))))).......)).. ( -39.32) >DroSec_CAF1 14375 120 + 1 AGCCACUAUCUCAAAGAUGGCCCAUUUGGGCGGCUCUCGCUGUCGUUGUUGUUGCCGUAUUUGCCCGUCUUGACGGCACUUAAAUUGCAUUAUUAUAUACGGCUCGACAUUUAUGUGCAA ((((..((((.....))))((((....))))))))...((......(((((..(((((((.(((((((....))))..........))).......))))))).))))).......)).. ( -35.82) >DroSim_CAF1 14517 120 + 1 AGCCACUAUCUCAAAGAUGGCCCAUUUGGGCGGCUCGCGCUGUCGUUGUUGUUGCCGUAUUUGCCCGUCUUGACGGCACUUAAAUUGCAUUAUUAUAUACGGCUCGACAUUUAUGUGCAA ((((..((((.....))))((((....)))))))).((((......(((((..(((((((.(((((((....))))..........))).......))))))).))))).....)))).. ( -39.90) >consensus AGCCACUAUCUCAAAGAUGGCCCAUUUGGGCGGCUCUCGCUGUCGUUGUUGUUGCCGUAUUUGCCCGUCUUGACGGCACUUAAAUUGCAUUAUUAUAUACGGCUCGACAUUUAUGUGCAA ((((..((((.....))))((((....))))))))...((......(((((..(((((((.(((.(((....))))))....(((......)))..))))))).))))).......)).. (-35.52 = -35.52 + -0.00)



| Location | 14,836,177 – 14,836,296 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -33.27 |
| Energy contribution | -33.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14836177 119 - 27905053 UUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUAAGUGCCGUCAAGACG-GCAAAUACGGCAACAACAACGACAGCGAGAGCCGCCCAAAUGGGCCAUCUUUGAGAUAGUGGCU ..((.(((...((((..(((((((....(((......)))..((((((....)))-))).))))))).....(((.((..((....)).((((....))))..)).))).))))))))). ( -36.90) >DroSec_CAF1 14375 120 - 1 UUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUAAGUGCCGUCAAGACGGGCAAAUACGGCAACAACAACGACAGCGAGAGCCGCCCAAAUGGGCCAUCUUUGAGAUAGUGGCU ..((.(((...((((..(((((((....(((......)))..((((((....)).)))).))))))).....(((.((..((....)).((((....))))..)).))).))))))))). ( -33.10) >DroSim_CAF1 14517 120 - 1 UUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUAAGUGCCGUCAAGACGGGCAAAUACGGCAACAACAACGACAGCGCGAGCCGCCCAAAUGGGCCAUCUUUGAGAUAGUGGCU ..((.(((...((((..(((((((....(((......)))..((((((....)).)))).))))))).....(((.((..((....)).((((....))))..)).))).))))))))). ( -33.60) >consensus UUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUAAGUGCCGUCAAGACGGGCAAAUACGGCAACAACAACGACAGCGAGAGCCGCCCAAAUGGGCCAUCUUUGAGAUAGUGGCU ..((.(((...((((..(((((((....(((......)))..((((((....))).))).))))))).....(((.((..((....)).((((....))))..)).))).))))))))). (-33.27 = -33.27 + 0.00)



| Location | 14,836,217 – 14,836,336 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -24.15 |
| Energy contribution | -24.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14836217 119 - 27905053 GAAGCGAAGCAAAUGUGUCCAACUUUGAACGAUAAAAUAUUUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUAAGUGCCGUCAAGACG-GCAAAUACGGCAACAACAACGACA ........((((((((....................)))))))).......(((((.(((((((....(((......)))..((((((....)))-))).)))))))........))))) ( -27.95) >DroSec_CAF1 14415 120 - 1 GAAGCGAAGCAAAUGUGUCCAACUUUGAACGAUAAAAUAUUUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUAAGUGCCGUCAAGACGGGCAAAUACGGCAACAACAACGACA ........((((((((....................)))))))).......(((((.(((((((....(((......)))..((((((....)).)))).)))))))........))))) ( -24.15) >DroSim_CAF1 14557 120 - 1 GAAGCGAAGCAAAUGUGUCCAACUUUGAACGAUAAAAUAUUUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUAAGUGCCGUCAAGACGGGCAAAUACGGCAACAACAACGACA ........((((((((....................)))))))).......(((((.(((((((....(((......)))..((((((....)).)))).)))))))........))))) ( -24.15) >consensus GAAGCGAAGCAAAUGUGUCCAACUUUGAACGAUAAAAUAUUUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUAAGUGCCGUCAAGACGGGCAAAUACGGCAACAACAACGACA ........((((((((....................)))))))).......(((((.(((((((....(((......)))..((((((....))).))).)))))))........))))) (-24.15 = -24.15 + 0.00)



| Location | 14,836,256 – 14,836,376 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -24.55 |
| Energy contribution | -24.55 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14836256 120 - 27905053 AGCCAAAUUCUUUUUUGCGGCUCUUUUUCCAUCAAUGGACGAAGCGAAGCAAAUGUGUCCAACUUUGAACGAUAAAAUAUUUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUA .((...(((.((...(((((((((((.((((....)))).))).....((((((((....................))))))))...........))))))))...)).)))))...... ( -25.05) >DroSec_CAF1 14455 117 - 1 --CCAAAUUCUU-UUUGCGGCUCUUUUUCCAUCAAUGGACGAAGCGAAGCAAAUGUGUCCAACUUUGAACGAUAAAAUAUUUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUA --..........-..(((((((((((.((((....)))).))).....((((((((....................))))))))...........))))))))................. ( -24.55) >DroSim_CAF1 14597 117 - 1 --CCAAAUUCUU-UUUGCGGCUCUUUUUCCAUCAAUGGACGAAGCGAAGCAAAUGUGUCCAACUUUGAACGAUAAAAUAUUUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUA --..........-..(((((((((((.((((....)))).))).....((((((((....................))))))))...........))))))))................. ( -24.55) >consensus __CCAAAUUCUU_UUUGCGGCUCUUUUUCCAUCAAUGGACGAAGCGAAGCAAAUGUGUCCAACUUUGAACGAUAAAAUAUUUGCACAUAAAUGUCGAGCCGUAUAUAAUAAUGCAAUUUA ...............(((((((((((.((((....)))).))).....((((((((....................))))))))...........))))))))................. (-24.55 = -24.55 + 0.00)

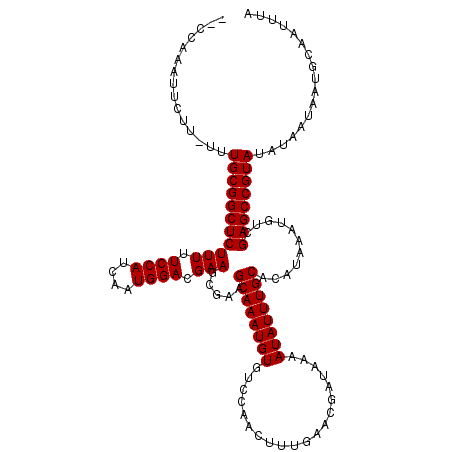

| Location | 14,836,336 – 14,836,456 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.96 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.26 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14836336 120 - 27905053 UUUAGGCCAUAAAGAAAUGCCUGAAUCCGACAAUGAGUCUGGGCUUUGGCUCUUUCUCCUUUCUUUUGAGAGCAGCAGCCAGCCAAAUUCUUUUUUGCGGCUCUUUUUCCAUCAAUGGAC (((((((.((......))))))))).(((.(((.((((...((((..(((((((((((.........))))).)).))))))))..))))....)))))).......((((....)))). ( -32.40) >DroSec_CAF1 14535 115 - 1 UUUAGGCCAUAACGAAAUGCCUGAAUCCGACAAUGAGUCUGGGCCUUGGCUCUUUCUCCUUUCUUUUGAGAGCAGCAG----CCAAAUUCUU-UUUGCGGCUCUUUUUCCAUCAAUGGAC (((((((.((......)))))))))((((((.....))).((((((((((((((((((.........))))).)).))----))))......-.....))))).............))). ( -29.60) >DroSim_CAF1 14677 111 - 1 CUUAGGCCAUA----AAUGCCUGAAUCCGACAAUGAGUCUGGGUCUUGGCUCUUUCUCCUUUCUUUUGAGAGCAGCAG----CCAAAUUCUU-UUUGCGGCUCUUUUUCCAUCAAUGGAC .((((((....----...))))))..........(((((.(((..(((((((((((((.........))))).)).))----))))..))).-.....)))))....((((....)))). ( -28.10) >consensus UUUAGGCCAUAA_GAAAUGCCUGAAUCCGACAAUGAGUCUGGGCCUUGGCUCUUUCUCCUUUCUUUUGAGAGCAGCAG____CCAAAUUCUU_UUUGCGGCUCUUUUUCCAUCAAUGGAC .((((((.((......)))))))).((((.....(((...((((....))))...))).........((((((.((((................)))).))))))..........)))). (-27.48 = -27.26 + -0.22)



| Location | 14,836,416 – 14,836,527 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -23.57 |
| Energy contribution | -25.57 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14836416 111 - 27905053 CCCACCCAC---------ACCAUGGAAAAUCUACAUAAAAACACACUUGAGCUAGAUUUCCAAUGGCCAAAGUGAGGCAGUUUAGGCCAUAAAGAAAUGCCUGAAUCCGACAAUGAGUCU ..(((....---------.(((((((((.(((((.(((........))).).))))))))).)))).....))).((..((((((((.((......))))))))))))(((.....))). ( -24.50) >DroSec_CAF1 14610 120 - 1 CCCACCCACCAUGGAUUUUCCAUGGAAAAUCUACAUAAAAACACACUUGAGCUAGAUUUCCUAUGGCCAAAGUGAGGCAGUUUAGGCCAUAACGAAAUGCCUGAAUCCGACAAUGAGUCU ........((((((.....))))))..................(((((..((((.........))))..))))).((..((((((((.((......))))))))))))(((.....))). ( -31.00) >DroSim_CAF1 14752 116 - 1 CCCACCCACCAUGGAUUUUCCAUGGAAAAUCUACAUAAAAACACACUUGAGCUAGAUUUCCAAUGGCCAAAGUGAGGCAGCUUAGGCCAUA----AAUGCCUGAAUCCGACAAUGAGUCU ............((((((..((((((((.(((((.(((........))).).))))))))).((((((.((((......)))).)))))).----.)))...))))))(((.....))). ( -29.40) >consensus CCCACCCACCAUGGAUUUUCCAUGGAAAAUCUACAUAAAAACACACUUGAGCUAGAUUUCCAAUGGCCAAAGUGAGGCAGUUUAGGCCAUAA_GAAAUGCCUGAAUCCGACAAUGAGUCU ........((((((.....))))))..................(((((..((((.........))))..))))).((..((((((((.((......))))))))))))(((.....))). (-23.57 = -25.57 + 2.00)

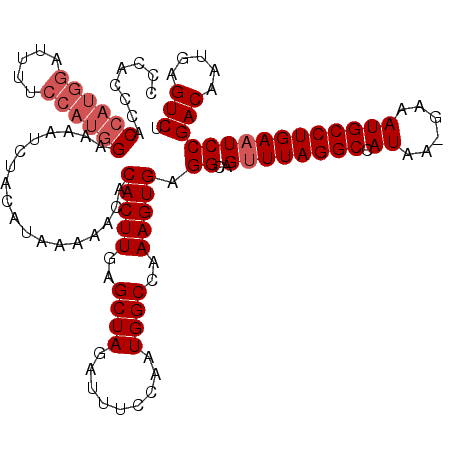

| Location | 14,836,496 – 14,836,607 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -21.82 |
| Energy contribution | -24.27 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14836496 111 - 27905053 UAACUGUGGAUGGAGGAGCUAGCUUACAUUGUGCCCCAACUCAGCAGUUUUUCAUCCCCCAAAAAAUUUCGAAAAUUACACCCACCCAC---------ACCAUGGAAAAUCUACAUAAAA ....(((((((.((((((((.(((....(((.....)))...)))))))))))................................(((.---------....)))...)))))))..... ( -18.10) >DroSec_CAF1 14690 119 - 1 UAACUGUGGAUGGGGGAGC-AACUUACAUGGUGCCCCAACUCAGCAGUUUUUCAUCCCCCAAAAAAAUUCGUAAAUUACACCCACCCACCAUGGAUUUUCCAUGGAAAAUCUACAUAAAA ....(((((((((((((((-(.((.....)))))...((((....)))).....))))))............................((((((.....))))))...)))))))..... ( -32.10) >DroSim_CAF1 14828 119 - 1 UAACUGUGGAGGGGGGAGC-AGCUUACAUGGUGCCCCAACUCAGCAGUUUUUCAUCCCCCAAAAAAUUUCGAAAAUUACACCCACCCACCAUGGAUUUUCCAUGGAAAAUCUACAUAAAA .((((((.(((..(((.((-(.(......).))))))..))).)))))).......................................((((((.....))))))............... ( -33.80) >consensus UAACUGUGGAUGGGGGAGC_AGCUUACAUGGUGCCCCAACUCAGCAGUUUUUCAUCCCCCAAAAAAUUUCGAAAAUUACACCCACCCACCAUGGAUUUUCCAUGGAAAAUCUACAUAAAA ....(((((((((((((...((((.....(((......)))....)))).....))))))............................((((((.....))))))...)))))))..... (-21.82 = -24.27 + 2.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:50 2006