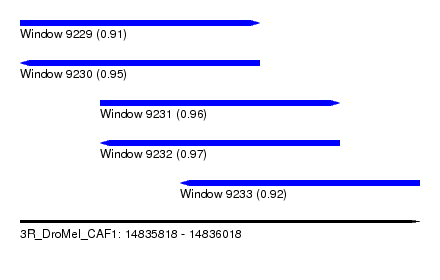
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,835,818 – 14,836,018 |
| Length | 200 |
| Max. P | 0.970558 |

| Location | 14,835,818 – 14,835,938 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.93 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -27.73 |
| Energy contribution | -28.73 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14835818 120 + 27905053 CGACAACUUGGAUCCCUGUUCAAUAGGUUGCCAAACUUGUUUUGGCAAAAACAACGCCGGUUGUCAAUCCCAGUCGACUGUCGACUGUCGACUGUCGCCAGCUAUCCUCAUUUUUUGUUU .(((((((.((...(((((...)))))((((((((.....))))))))........))))))))).....((((((((........)))))))).......................... ( -35.80) >DroSec_CAF1 14034 101 + 1 CGACAACUUGGAUCCCUGUUCAAUAGGUUGCCAAACUUGUUUUGGCAAAAACAACGCCGGUUGUCAAUCCCAGUCGAC--------------UGUCGCCAGCUAUCCUGGUUUU-----U .(((((((.((...(((((...)))))((((((((.....))))))))........))))))))).....(((....)--------------))..(((((.....)))))...-----. ( -29.10) >DroSim_CAF1 14169 108 + 1 CGACAACUUGGAUCCCUGUUCAAUAGGUUGCCAAACUUGUUUUGGCAAAAACAACGCCGGUUGUCAAUCCCAGUCGACUGU-------CGACUGUCGCCAGCUAUCCUGGUUUU-----U .(((((((.((...(((((...)))))((((((((.....))))))))........))))))))).....((((((.....-------))))))..(((((.....)))))...-----. ( -36.30) >consensus CGACAACUUGGAUCCCUGUUCAAUAGGUUGCCAAACUUGUUUUGGCAAAAACAACGCCGGUUGUCAAUCCCAGUCGACUGU_______CGACUGUCGCCAGCUAUCCUGGUUUU_____U .(((((((.((...(((((...)))))((((((((.....))))))))........))))))))).......(.((((.((((.....)))).)))).)..................... (-27.73 = -28.73 + 1.00)

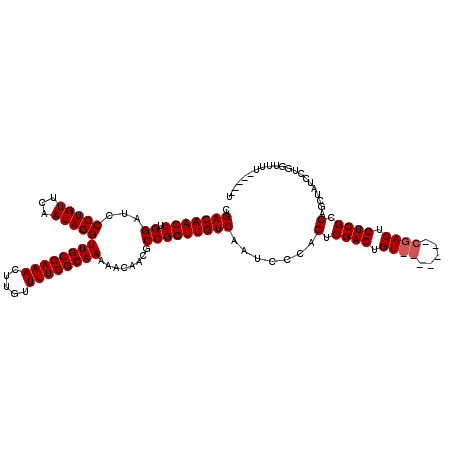
| Location | 14,835,818 – 14,835,938 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.93 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -32.07 |
| Energy contribution | -33.07 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14835818 120 - 27905053 AAACAAAAAAUGAGGAUAGCUGGCGACAGUCGACAGUCGACAGUCGACUGGGAUUGACAACCGGCGUUGUUUUUGCCAAAACAAGUUUGGCAACCUAUUGAACAGGGAUCCAAGUUGUCG ..............(((((((.....((((((((........))))))))(((((((((((....)))))).((((((((.....))))))))(((.......)))))))).))))))). ( -40.40) >DroSec_CAF1 14034 101 - 1 A-----AAAACCAGGAUAGCUGGCGACA--------------GUCGACUGGGAUUGACAACCGGCGUUGUUUUUGCCAAAACAAGUUUGGCAACCUAUUGAACAGGGAUCCAAGUUGUCG .-----....((((....((((....))--------------))...))))....((((((.(((.((((((((((((((.....))))))))......)))))).)..))..)))))). ( -32.70) >DroSim_CAF1 14169 108 - 1 A-----AAAACCAGGAUAGCUGGCGACAGUCG-------ACAGUCGACUGGGAUUGACAACCGGCGUUGUUUUUGCCAAAACAAGUUUGGCAACCUAUUGAACAGGGAUCCAAGUUGUCG .-----........(((((((.....((((((-------.....))))))(((((((((((....)))))).((((((((.....))))))))(((.......)))))))).))))))). ( -36.40) >consensus A_____AAAACCAGGAUAGCUGGCGACAGUCG_______ACAGUCGACUGGGAUUGACAACCGGCGUUGUUUUUGCCAAAACAAGUUUGGCAACCUAUUGAACAGGGAUCCAAGUUGUCG ..............(((((((((((((.((((.....)))).)))).)).(((((((((((....)))))).((((((((.....))))))))(((.......)))))))).))))))). (-32.07 = -33.07 + 1.00)



| Location | 14,835,858 – 14,835,978 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.21 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -21.83 |
| Energy contribution | -23.50 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14835858 120 + 27905053 UUUGGCAAAAACAACGCCGGUUGUCAAUCCCAGUCGACUGUCGACUGUCGACUGUCGCCAGCUAUCCUCAUUUUUUGUUUUUUUCUCAUCUUUGUUUUUGCCAGCUGCAAUCGACUUUUA .(((((((((((((.((.(((.(.((.((.((((((.....))))))..)).)).)))).))......((.....))..............)))))))))))))................ ( -31.60) >DroSec_CAF1 14074 101 + 1 UUUGGCAAAAACAACGCCGGUUGUCAAUCCCAGUCGAC--------------UGUCGCCAGCUAUCCUGGUUUU-----UUUUUCUCAUUUUUGUUUUUGCCAGCUCCAAUCGACUUUUA .(((((((((((((.(.((((((.(.......).))))--------------)).)(((((.....)))))...-----............)))))))))))))................ ( -27.40) >DroSim_CAF1 14209 108 + 1 UUUGGCAAAAACAACGCCGGUUGUCAAUCCCAGUCGACUGU-------CGACUGUCGCCAGCUAUCCUGGUUUU-----UUUUUCACAUUUUUGUUUUUGCCAGCUCCAAUCGACUUUUA .(((((((((((((.(.((((((.((.((......)).)).-------)))))).)(((((.....)))))...-----............)))))))))))))................ ( -30.10) >consensus UUUGGCAAAAACAACGCCGGUUGUCAAUCCCAGUCGACUGU_______CGACUGUCGCCAGCUAUCCUGGUUUU_____UUUUUCUCAUUUUUGUUUUUGCCAGCUCCAAUCGACUUUUA .(((((((((((((....(..((.......))..)(((.((((.....)))).)))(((((.....)))))....................)))))))))))))................ (-21.83 = -23.50 + 1.67)

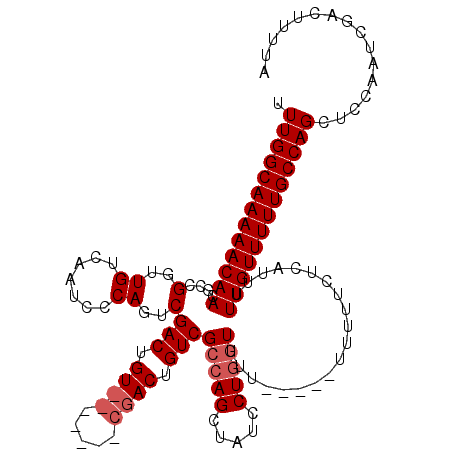

| Location | 14,835,858 – 14,835,978 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.21 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -25.75 |
| Energy contribution | -26.87 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14835858 120 - 27905053 UAAAAGUCGAUUGCAGCUGGCAAAAACAAAGAUGAGAAAAAAACAAAAAAUGAGGAUAGCUGGCGACAGUCGACAGUCGACAGUCGACUGGGAUUGACAACCGGCGUUGUUUUUGCCAAA .................((((((((((((..............((.....))......(((((...(((((..((((((.....)))))).)))))....))))).)))))))))))).. ( -37.20) >DroSec_CAF1 14074 101 - 1 UAAAAGUCGAUUGGAGCUGGCAAAAACAAAAAUGAGAAAAA-----AAAACCAGGAUAGCUGGCGACA--------------GUCGACUGGGAUUGACAACCGGCGUUGUUUUUGCCAAA ....(((((((((..((..((....................-----............))..))..))--------------)))))))((.(..((((((....))))))..).))... ( -31.75) >DroSim_CAF1 14209 108 - 1 UAAAAGUCGAUUGGAGCUGGCAAAAACAAAAAUGUGAAAAA-----AAAACCAGGAUAGCUGGCGACAGUCG-------ACAGUCGACUGGGAUUGACAACCGGCGUUGUUUUUGCCAAA .....((((((((..((..((.............(....).-----............))..))..))))))-------)).......(((.(..((((((....))))))..).))).. ( -32.27) >consensus UAAAAGUCGAUUGGAGCUGGCAAAAACAAAAAUGAGAAAAA_____AAAACCAGGAUAGCUGGCGACAGUCG_______ACAGUCGACUGGGAUUGACAACCGGCGUUGUUUUUGCCAAA .................((((((((((((.....................((((.....))))((((.((((.....)))).)))).((((.........))))..)))))))))))).. (-25.75 = -26.87 + 1.11)



| Location | 14,835,898 – 14,836,018 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -19.65 |
| Energy contribution | -19.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14835898 120 - 27905053 UAAAAGUCAUUUUAUUAAGAAGCAGUCGACAGAAUGGAGCUAAAAGUCGAUUGCAGCUGGCAAAAACAAAGAUGAGAAAAAAACAAAAAAUGAGGAUAGCUGGCGACAGUCGACAGUCGA .........................(((((((.......))....((((((((..((..((.....((....)).........((.....))......))..))..)))))))).))))) ( -25.30) >DroSec_CAF1 14112 103 - 1 UAAAAGUCAUUUUAUUAAGAAGCAGUCGACAGAAUGGAGCUAAAAGUCGAUUGGAGCUGGCAAAAACAAAAAUGAGAAAAA-----AAAACCAGGAUAGCUGGCGACA------------ .....(((..............((((((((...............))))))))..((..((....................-----............))..))))).------------ ( -20.11) >DroSim_CAF1 14249 108 - 1 UAAAAGUCAUUUUAUUAAGAAGCAGUCGACAGAAUGGAGCUAAAAGUCGAUUGGAGCUGGCAAAAACAAAAAUGUGAAAAA-----AAAACCAGGAUAGCUGGCGACAGUCG-------A .....(((..............((((((((...............))))))))..((..((.............(....).-----............))..))))).....-------. ( -20.53) >consensus UAAAAGUCAUUUUAUUAAGAAGCAGUCGACAGAAUGGAGCUAAAAGUCGAUUGGAGCUGGCAAAAACAAAAAUGAGAAAAA_____AAAACCAGGAUAGCUGGCGACAGUCG_______A .....(((..............((((((((...............))))))))..((..((.....................................))..)))))............. (-19.65 = -19.65 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:44 2006