| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,104,821 – 2,104,931 |
| Length | 110 |
| Max. P | 0.883822 |

| Location | 2,104,821 – 2,104,931 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
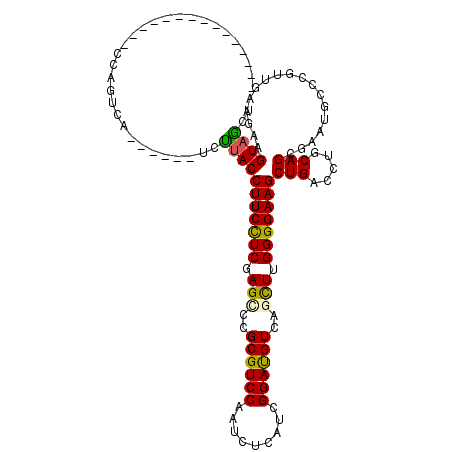
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -21.15 |
| Energy contribution | -20.85 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
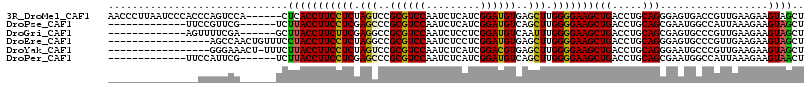
>3R_DroMel_CAF1 2104821 110 - 27905053 AACCCUUAAUCCCACCCAGUCCA------CUCACCUUCCUCUAGUCCGCGUCCAAUCUCAUCGGAUGUGAGCUUGGGGAAGCUGACCUGCAGGGAGUGACCGUUGAAGAAGUAGCU (((..(((.((((...(((....------.(((.(((((((.((..(((((((.........)))))))..)).))))))).))).)))..)))).)))..)))............ ( -34.40) >DroPse_CAF1 223918 97 - 1 -------------UUCCGUUCG------UCUUACCUUCCUCGAGCCCGCGUCCAAUCUCAUCGGAUGUCAGCUUGGGGAAGCUGACCUGCAGCGAAUGGCCAUUAAAGAAGUAGCU -------------..((((((.------.......((((((((((..((((((.........))))))..))))))))))((((.....))))))))))................. ( -35.50) >DroGri_CAF1 255877 97 - 1 -------------AGUUUUCGA------GCUUACCUUCUUCGAGGCCGCGUCCAAUCUCCUCGGAUGUCAAUUUGGGGAAGCUGACCUGCAGCGAGUGCCCGUUGAAGAAGUAGCU -------------........(------(((...(((((((((((.(((........(((((((((....))))))))).((((.....))))..))).)).))))))))).)))) ( -35.70) >DroEre_CAF1 249893 99 - 1 -----------------AGCCAACUGUUUCCUACCUUCCUCUAGGCCGCGUCCAAUCUCCUCGGAUGUGAGCUUGGGGAAGCUGACCUGCAGGGAGUGCCCGUUGAAGAAGUAGCU -----------------(((..(((.((((..(((((((.(((((((((((((.........))))))).)))))))))))..........(((....))))).)))).))).))) ( -34.50) >DroYak_CAF1 251370 98 - 1 -----------------GGGAAACU-UUUCUUACCUUCCUCUAGUCCGCGUCCAAUCUCAUCGGACGUGAGCUUGGGGAAGCUGACCUGCAGGGAAUGCCCGUUGAAGAAGUAGCU -----------------((....))-((((((..(((((((.((..(((((((.........)))))))..)).)))))))..........(((....)))....))))))..... ( -34.20) >DroPer_CAF1 225858 97 - 1 -------------UUCCAUUCG------UCUUACCUUCCUCGAGCCCGCGUCCAAUCUCAUCGGAUGUCAGCUUGGGGAAGCUGACCUGCAGCGAAUGGCCAUUAAAGAAGUAACU -------------..((((((.------.......((((((((((..((((((.........))))))..))))))))))((((.....))))))))))................. ( -35.90) >consensus _______________CCAGUCA______UCUUACCUUCCUCGAGCCCGCGUCCAAUCUCAUCGGAUGUCAGCUUGGGGAAGCUGACCUGCAGCGAAUGCCCGUUGAAGAAGUAGCU ..............................(((((((((((.(((..((((((.........))))))..))).)))))))(((.....)))..................)))).. (-21.15 = -20.85 + -0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:11 2006