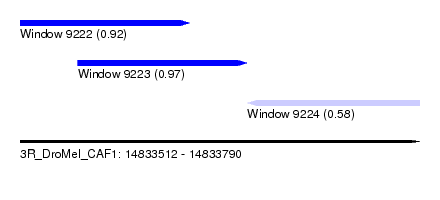
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,833,512 – 14,833,790 |
| Length | 278 |
| Max. P | 0.973833 |

| Location | 14,833,512 – 14,833,630 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -35.89 |
| Energy contribution | -35.23 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14833512 118 + 27905053 CCUUCAUGUCCACAAACUGGUCACCAGUGCCGAGAGAAUAAAAUAUGCAUGUUGCAGGAGCACC--AGCUUCGUGGCAUGUGGCCGGGUUAUUAAAGUUAAUUACAAUUCUUCCUGGCAU .....(((.(((....((((((((..((((((.............(((.....)))(((((...--.))))).))))))))))))))((.((((....)))).)).........)))))) ( -33.10) >DroSec_CAF1 11699 118 + 1 CCUUCCUGUCCACAAACUGGCCACCACUGCCGAGAGAAAUAAAUAUGCAUGUUGCAGGAGCACC--AGCUCCGUGGCAUGUGGCCGGGUUAUUAAAGUUAAUUACAAUUCUUCCUGGCAU ......((.(((....((((((((...(((((.............(((.....)))(((((...--.))))).))))).))))))))((.((((....)))).)).........))))). ( -36.50) >DroSim_CAF1 11849 120 + 1 CCUUCCUGUCCACAAACUGGCCACCACUGCCAAGAGAAUUAAAUAUGCAUGUUGCAGGAGCUCCGUGGCUCCGUGGCAUGUGGCCGGGUUAUUAAAGUUAAUUACAAUUCUUCCUGGCAU ......((.(((....((((((((...(((((.............(((.....)))((((((....)))))).))))).))))))))((.((((....)))).)).........))))). ( -37.80) >consensus CCUUCCUGUCCACAAACUGGCCACCACUGCCGAGAGAAUUAAAUAUGCAUGUUGCAGGAGCACC__AGCUCCGUGGCAUGUGGCCGGGUUAUUAAAGUUAAUUACAAUUCUUCCUGGCAU ......((.(((....((((((((...(((((.............(((.....)))(((((......))))).))))).))))))))((.((((....)))).)).........))))). (-35.89 = -35.23 + -0.66)



| Location | 14,833,552 – 14,833,670 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -29.71 |
| Energy contribution | -29.49 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14833552 118 + 27905053 AAAUAUGCAUGUUGCAGGAGCACC--AGCUUCGUGGCAUGUGGCCGGGUUAUUAAAGUUAAUUACAAUUCUUCCUGGCAUUCAUUUUUUCUCGUAAUGAUAAUUCUGAUUUUUUUUCACC ......((((((..(.(((((...--.))))))..)))))).((((((........((.....)).......))))))..(((((.........)))))..................... ( -28.46) >DroSec_CAF1 11739 115 + 1 AAAUAUGCAUGUUGCAGGAGCACC--AGCUCCGUGGCAUGUGGCCGGGUUAUUAAAGUUAAUUACAAUUCUUCCUGGCAUUCAUUUUUUCACGAAACGAUAAUUCUGAUUUUUUUUU--- ......((((((..(.(((((...--.))))))..)))))).((((((........((.....)).......))))))..........(((.(((.......)))))).........--- ( -30.96) >DroSim_CAF1 11889 117 + 1 AAAUAUGCAUGUUGCAGGAGCUCCGUGGCUCCGUGGCAUGUGGCCGGGUUAUUAAAGUUAAUUACAAUUCUUCCUGGCAUUCAUUUUUUCUCGUAACGAUAAUUCUGAUUUUUUUUU--- ......((((((..(.((((((....)))))))..)))))).((((((........((.....)).......)))))).......................................--- ( -30.76) >consensus AAAUAUGCAUGUUGCAGGAGCACC__AGCUCCGUGGCAUGUGGCCGGGUUAUUAAAGUUAAUUACAAUUCUUCCUGGCAUUCAUUUUUUCUCGUAACGAUAAUUCUGAUUUUUUUUU___ ......((((((..(.(((((......))))))..)))))).((((((........((.....)).......)))))).......................................... (-29.71 = -29.49 + -0.22)

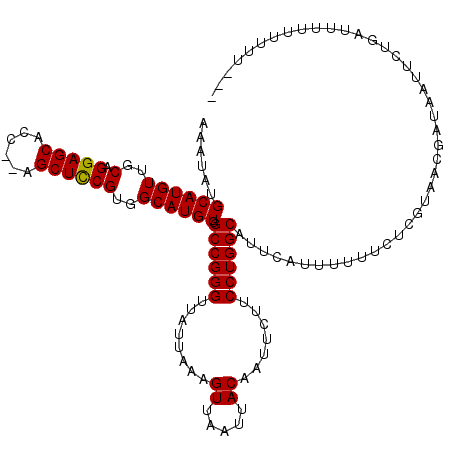

| Location | 14,833,670 – 14,833,790 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.93 |
| Mean single sequence MFE | -25.19 |
| Consensus MFE | -22.32 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14833670 120 - 27905053 GCCUCCGAUUUUGCAGUCUGUAAACAAAGGAAGCAAAAACAAGGCGAUGCCGCAAAUAAAUCAACGCGAAAAUUAAUAGAGUGGAAAAAUCGGCACAGAGCAAGGACACAGCACUUAACU ((((((((((((.((.(((((.....................(((...)))((............))........))))).))..))))))))...)).))................... ( -21.70) >DroSec_CAF1 11854 115 - 1 GCCUCUGAUUUUGCAGUCUGUAAACAAAGGAAGCAAAAACAAGGCGAUGCCGCAAAUAAAUCAACACGAAAAUUAAUAGAGUGGAAAAAUCGCCACAGAGCAAGGACACAGCAUU----- ((.((((.((((((..(((........)))..))))))....((((((.((((...........................))))....)))))).))))))..............----- ( -28.23) >DroSim_CAF1 12006 115 - 1 GCCUCCGAUUUUGCAGUCUGUAAACAAAGGAAGCAAAAACAAGGCGAUGCCGCAAAUAAAUCAACGCGAAAAUUAAUAGAGUGGAAAAAUCGCCACAGAGCAAGGACACAGCAUU----- ((.((((.((((((..(((........)))..)))))).)..((((((.((((...........................))))....)))))).........)))....))...----- ( -25.63) >consensus GCCUCCGAUUUUGCAGUCUGUAAACAAAGGAAGCAAAAACAAGGCGAUGCCGCAAAUAAAUCAACGCGAAAAUUAAUAGAGUGGAAAAAUCGCCACAGAGCAAGGACACAGCAUU_____ ((.((((.((((((..(((........)))..)))))).)..((((((.((((...........................))))....)))))).........)))....))........ (-22.32 = -22.43 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:36 2006