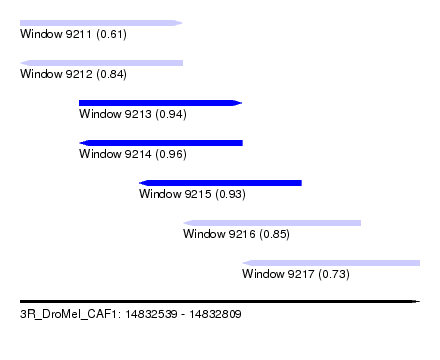
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,832,539 – 14,832,809 |
| Length | 270 |
| Max. P | 0.959602 |

| Location | 14,832,539 – 14,832,649 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -28.72 |
| Energy contribution | -29.63 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14832539 110 + 27905053 GAUUUGUGGCAAUCUCUGUGGGCAACGAUUGCUAUCUGGCCCAGCCUACAUGGCAUGGCUUUGUGUACAUAAGUUUAUGU----------ACACAGACAUUCCUGCGCACAUGACUCACA ((..(((((((.......(((((...(((....)))..)))))(((.....)))..((.((((((((((((....)))))----------)))))))....))))).))))....))... ( -36.00) >DroSec_CAF1 10713 120 + 1 GAUUUGUGUCAAUCUCUGUGGGCAACGAUUGCUAUCUGUCGCAGCCUACAUGGCAUGGCUUUGUGUACAUAGGUUUAUGUACAGAUGUGUACACAGACAUUCCUGCGCACAUGACUCACA ....((.((((.....(((((((..((((........))))..)))))))(((((.((.((((((((((((.((......))...))))))))))))....))))).))..)))).)).. ( -39.80) >DroSim_CAF1 10875 120 + 1 GAUUUGUGGCAAUCUCUGUGGGCAACGAUUGCUAUCUGUCGCAGCCUACAUGGCAUGGCUUUGUGUACAUAGGUUUAUGUACAGAUGUGUACACAGACAUUCCUGCGCACAUGACUCACA ((.(..((((......(((((((..((((........))))..)))))))..(((.((.((((((((((((.((......))...))))))))))))....))))))).))..).))... ( -41.60) >consensus GAUUUGUGGCAAUCUCUGUGGGCAACGAUUGCUAUCUGUCGCAGCCUACAUGGCAUGGCUUUGUGUACAUAGGUUUAUGUACAGAUGUGUACACAGACAUUCCUGCGCACAUGACUCACA ....((((.....((.(((((((..((((........))))..))))))).))((((....((((((.....((((.((((((....)))))).)))).....))))))))))...)))) (-28.72 = -29.63 + 0.92)


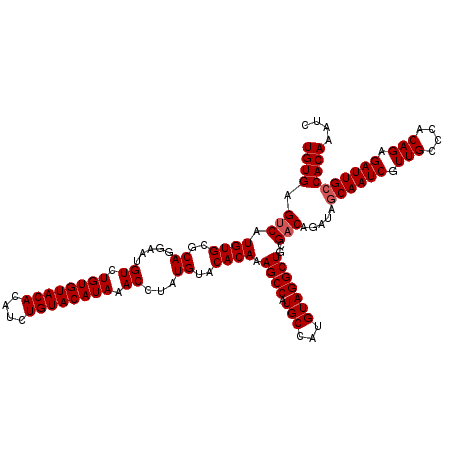
| Location | 14,832,539 – 14,832,649 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -30.48 |
| Energy contribution | -31.40 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14832539 110 - 27905053 UGUGAGUCAUGUGCGCAGGAAUGUCUGUGU----------ACAUAAACUUAUGUACACAAAGCCAUGCCAUGUAGGCUGGGCCAGAUAGCAAUCGUUGCCCACAGAGAUUGCCACAAAUC ((((((((.((((.((((((.(((((((((----------(((((....)))))))))...(((..(((.....)))..))).)))))....)).)))).))))..))))..)))).... ( -37.80) >DroSec_CAF1 10713 120 - 1 UGUGAGUCAUGUGCGCAGGAAUGUCUGUGUACACAUCUGUACAUAAACCUAUGUACACAAAGCCAUGCCAUGUAGGCUGCGACAGAUAGCAAUCGUUGCCCACAGAGAUUGACACAAAUC (((.((((.((((((((((....))))))))))..((((((((((....)))))))....((((.(((...)))))))(((((.(((....))))))))....))))))).)))...... ( -37.70) >DroSim_CAF1 10875 120 - 1 UGUGAGUCAUGUGCGCAGGAAUGUCUGUGUACACAUCUGUACAUAAACCUAUGUACACAAAGCCAUGCCAUGUAGGCUGCGACAGAUAGCAAUCGUUGCCCACAGAGAUUGCCACAAAUC ((((.(((.((((((((((....))))))))))....((((((((....))))))))....((...(((.....))).))))).....((((((.(((....))).)))))))))).... ( -40.60) >consensus UGUGAGUCAUGUGCGCAGGAAUGUCUGUGUACACAUCUGUACAUAAACCUAUGUACACAAAGCCAUGCCAUGUAGGCUGCGACAGAUAGCAAUCGUUGCCCACAGAGAUUGCCACAAAUC ((((.(((.((((..((.....((.((((((((....)))))))).))...))..)))).((((.(((...)))))))..))).....((((((.(((....))).)))))))))).... (-30.48 = -31.40 + 0.92)


| Location | 14,832,579 – 14,832,689 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -35.05 |
| Energy contribution | -35.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14832579 110 + 27905053 CCAGCCUACAUGGCAUGGCUUUGUGUACAUAAGUUUAUGU----------ACACAGACAUUCCUGCGCACAUGACUCACAAAUAAUGUGCAAUGGCAGCCAUGACCAUGGUCGUCACCCA ........(((((((((((((((((((((((....)))))----------)))))))....((...((((((............))))))...))..)))))).)))))........... ( -40.20) >DroSec_CAF1 10753 120 + 1 GCAGCCUACAUGGCAUGGCUUUGUGUACAUAGGUUUAUGUACAGAUGUGUACACAGACAUUCCUGCGCACAUGACUCACAAAUAAUGUGCAAUGGCAGCCAUGACCAUGGUCGUCACCCA ........(((((((((((((((((((((((.((......))...))))))))))))....((...((((((............))))))...))..)))))).)))))........... ( -41.90) >DroSim_CAF1 10915 120 + 1 GCAGCCUACAUGGCAUGGCUUUGUGUACAUAGGUUUAUGUACAGAUGUGUACACAGACAUUCCUGCGCACAUGACUCACAAAUAAUGUGCAAUGGCAGCCAUGACCAUGGUCGUCACCCA ........(((((((((((((((((((((((.((......))...))))))))))))....((...((((((............))))))...))..)))))).)))))........... ( -41.90) >consensus GCAGCCUACAUGGCAUGGCUUUGUGUACAUAGGUUUAUGUACAGAUGUGUACACAGACAUUCCUGCGCACAUGACUCACAAAUAAUGUGCAAUGGCAGCCAUGACCAUGGUCGUCACCCA ........(((((((((((((..((((.....((((.((((((....)))))).)))).....)))((((((............)))))).)..).))))))).)))))........... (-35.05 = -35.30 + 0.25)

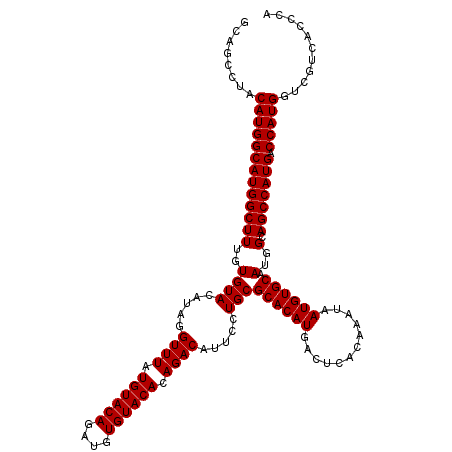

| Location | 14,832,579 – 14,832,689 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -37.40 |
| Energy contribution | -38.73 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14832579 110 - 27905053 UGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUUUGUGAGUCAUGUGCGCAGGAAUGUCUGUGU----------ACAUAAACUUAUGUACACAAAGCCAUGCCAUGUAGGCUGG ..(....)...(((((.(((((((.((..(((((((.(((....))).)))))))..))......(((((----------(((((....)))))))))).))))))))))))........ ( -42.10) >DroSec_CAF1 10753 120 - 1 UGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUUUGUGAGUCAUGUGCGCAGGAAUGUCUGUGUACACAUCUGUACAUAAACCUAUGUACACAAAGCCAUGCCAUGUAGGCUGC ..(....)...(((((.(((((((.......((((.....)))).....((((((((((....))))))))))....((((((((....))))))))...))))))))))))........ ( -43.10) >DroSim_CAF1 10915 120 - 1 UGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUUUGUGAGUCAUGUGCGCAGGAAUGUCUGUGUACACAUCUGUACAUAAACCUAUGUACACAAAGCCAUGCCAUGUAGGCUGC ..(....)...(((((.(((((((.......((((.....)))).....((((((((((....))))))))))....((((((((....))))))))...))))))))))))........ ( -43.10) >consensus UGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUUUGUGAGUCAUGUGCGCAGGAAUGUCUGUGUACACAUCUGUACAUAAACCUAUGUACACAAAGCCAUGCCAUGUAGGCUGC ..(....)...(((((.(((((((((....))).....((((((.....((((((((((....))))))))))......((((((....)))))))))))))))))))))))........ (-37.40 = -38.73 + 1.33)



| Location | 14,832,619 – 14,832,729 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -47.11 |
| Consensus MFE | -46.52 |
| Energy contribution | -46.74 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14832619 110 - 27905053 GGCCAAGGAUGCCAAGGGAAACCGGGGGCGUCGUCACGUCUGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUUUGUGAGUCAUGUGCGCAGGAAUGUCUGUGU---------- (((((..((((((..((....))...))))))(((((......)))))(((....))).)))))......((((((.(((....))).))))))(((((....)))))..---------- ( -45.50) >DroSec_CAF1 10793 120 - 1 GGCCAAGGGCGCCAAGGGAAUCCGGGGGCGUCGUCACGUCUGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUUUGUGAGUCAUGUGCGCAGGAAUGUCUGUGUACACAUCUGU (((((.(((((((..((....))...)))))((((((......)))))).)).)))))((((((.......((((.....))))))))))(((((((((....)))))))))........ ( -47.91) >DroSim_CAF1 10955 120 - 1 GGCCAAGGGCGCCAAGGGAAUCCGGGGGCGUCGUCACGUCUGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUUUGUGAGUCAUGUGCGCAGGAAUGUCUGUGUACACAUCUGU (((((.(((((((..((....))...)))))((((((......)))))).)).)))))((((((.......((((.....))))))))))(((((((((....)))))))))........ ( -47.91) >consensus GGCCAAGGGCGCCAAGGGAAUCCGGGGGCGUCGUCACGUCUGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUUUGUGAGUCAUGUGCGCAGGAAUGUCUGUGUACACAUCUGU (((((.(((((((..((....))...)))))((((((......)))))).)).)))))((((((.......((((.....))))))))))(((((((((....)))))))))........ (-46.52 = -46.74 + 0.23)


| Location | 14,832,649 – 14,832,769 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -42.13 |
| Consensus MFE | -42.57 |
| Energy contribution | -42.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14832649 120 - 27905053 CUUGGCAAAAAUUAUUAUUUUCCUCAAAAUACGCUCGGAGGGCCAAGGAUGCCAAGGGAAACCGGGGGCGUCGUCACGUCUGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUU ..(((((..............((((............))))((((..((((((..((....))...))))))(((((......)))))(((....))).)))))))))............ ( -44.10) >DroSec_CAF1 10833 120 - 1 CUUGGCAAAAAUUAUUAUUUUCCUCAAAAUACGCUCAGAGGGCCAAGGGCGCCAAGGGAAUCCGGGGGCGUCGUCACGUCUGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUU ..(((((..............((((............))))((((..((((((..((....))...))))))(((((......)))))(((....))).)))))))))............ ( -41.20) >DroSim_CAF1 10995 120 - 1 CUUGGCAAAAAUUAUUAUUUUCCUCAAAAUACGCUCGGAGGGCCAAGGGCGCCAAGGGAAUCCGGGGGCGUCGUCACGUCUGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUU ..(((((..............((((............))))((((..((((((..((....))...))))))(((((......)))))(((....))).)))))))))............ ( -41.10) >consensus CUUGGCAAAAAUUAUUAUUUUCCUCAAAAUACGCUCGGAGGGCCAAGGGCGCCAAGGGAAUCCGGGGGCGUCGUCACGUCUGGGUGACGACCAUGGUCAUGGCUGCCAUUGCACAUUAUU ..(((((..............((((............))))((((..((((((..((....))...))))))(((((......)))))(((....))).)))))))))............ (-42.57 = -42.13 + -0.44)



| Location | 14,832,689 – 14,832,809 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -40.43 |
| Consensus MFE | -40.87 |
| Energy contribution | -40.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14832689 120 - 27905053 CGGAAGCUGACAGCAGCCGUCGCCGGAAAUGGCAAAUUAACUUGGCAAAAAUUAUUAUUUUCCUCAAAAUACGCUCGGAGGGCCAAGGAUGCCAAGGGAAACCGGGGGCGUCGUCACGUC (((..((((....)))).....)))((..((((.......((((((.(((((....)))))((((............))))))))))((((((..((....))...))))))))))..)) ( -42.40) >DroSec_CAF1 10873 120 - 1 CGGAAGCUGACAGCAGCCGUCGCCGGAAAUGGCAAAUUAACUUGGCAAAAAUUAUUAUUUUCCUCAAAAUACGCUCAGAGGGCCAAGGGCGCCAAGGGAAUCCGGGGGCGUCGUCACGUC (((..((((....)))).....)))((..((((.......((((((.(((((....)))))((((............))))))))))((((((..((....))...))))))))))..)) ( -39.50) >DroSim_CAF1 11035 120 - 1 CGGAAGCUGACAGCAGCCGUCGCCGGAAAUGGCAAAUUAACUUGGCAAAAAUUAUUAUUUUCCUCAAAAUACGCUCGGAGGGCCAAGGGCGCCAAGGGAAUCCGGGGGCGUCGUCACGUC (((..((((....)))).....)))((..((((.......((((((.(((((....)))))((((............))))))))))((((((..((....))...))))))))))..)) ( -39.40) >consensus CGGAAGCUGACAGCAGCCGUCGCCGGAAAUGGCAAAUUAACUUGGCAAAAAUUAUUAUUUUCCUCAAAAUACGCUCGGAGGGCCAAGGGCGCCAAGGGAAUCCGGGGGCGUCGUCACGUC (((..((((....)))).....)))((..((((.......((((((.(((((....)))))((((............))))))))))((((((..((....))...))))))))))..)) (-40.87 = -40.43 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:29 2006