
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,828,444 – 14,828,601 |
| Length | 157 |
| Max. P | 0.772130 |

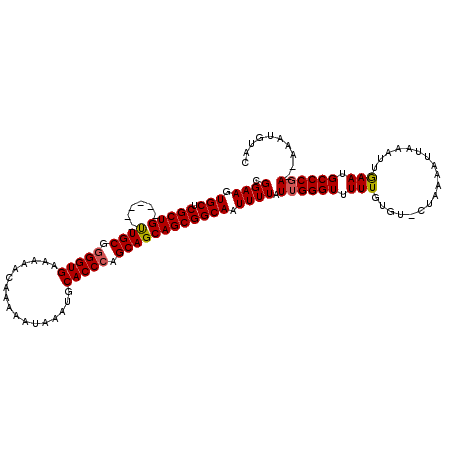
| Location | 14,828,444 – 14,828,561 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -37.14 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.67 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14828444 117 - 27905053 CGGAAGUGCUCGCUGCUGCUGCUGCGGGGUGAAAAACAAAAAUAAAUGCACCCAGCAGCAGCGGCAAUUUUAUUGGGUUUUUGUGU-CUAAAAUUAAAUUGAAUGCCCGA--AAAUGUUC (....).......((((((((((((.(((((.................))))).))))))))))))......((((((.((((...-............)))).))))))--........ ( -39.69) >DroSec_CAF1 6592 114 - 1 CGGAAGUGCUCGCUG------UUGCGGGGUGAAAAACAAAAAUAAAUGCACCAAGCAGCAGCGGCAAUUUUAUUGGGUUUUUGUGUUCUAAAAUUAAAUGAAAUGCCCCAAAAAAUGUAC .....((((..((((------((((..((((.................))))..))))))))((((.(((((((..(((((........)))))..))))))))))).........)))) ( -35.13) >DroSim_CAF1 6750 111 - 1 CGGAAGUGCUCGCUG------UUGCGGGGUGAAAAACAAAAAUAAAUGCACCCAGCAGCAGCGGCAAUUUUAUUGGGUUUUUGUGU-CUAAAAUUAAAUUGAAUGCCCGA--AAAUGUAC .((((.(((.(((((------((((.(((((.................))))).)))))))))))).)))).((((((.((((...-............)))).))))))--........ ( -36.59) >consensus CGGAAGUGCUCGCUG______UUGCGGGGUGAAAAACAAAAAUAAAUGCACCCAGCAGCAGCGGCAAUUUUAUUGGGUUUUUGUGU_CUAAAAUUAAAUUGAAUGCCCGA__AAAUGUAC .((((.(((.(((((......((((.(((((.................))))).)))))))))))).)))).((((((.(((..................))).)))))).......... (-26.44 = -26.67 + 0.23)


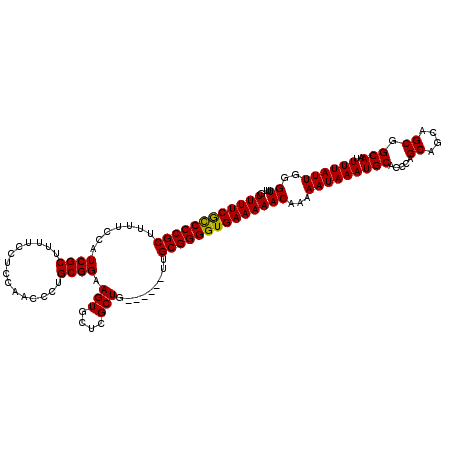
| Location | 14,828,481 – 14,828,601 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -30.77 |
| Energy contribution | -30.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14828481 120 - 27905053 GUUUUCGCCCCGCUUUUCCAUCGCUUUUCCUCCAACCCUGCGGAAGUGCUCGCUGCUGCUGCUGCGGGGUGAAAAACAAAAAUAAAUGCACCCAGCAGCAGCGGCAAUUUUAUUGGGUUU ......((((.((.(((((...((...............))))))).))....((((((((((((.(((((.................))))).))))))))))))........)))).. ( -44.19) >DroSec_CAF1 6632 114 - 1 GUUUUCAUCCCGCUUUUCCAUCGCUUUUCCUCCAACCCUGCGGAAGUGCUCGCUG------UUGCGGGGUGAAAAACAAAAAUAAAUGCACCAAGCAGCAGCGGCAAUUUUAUUGGGUUU .................................(((((.(..(((.(((.(((((------((((..((((.................))))..)))))))))))).)))..).))))). ( -35.43) >DroSim_CAF1 6787 114 - 1 GUUUUCGCCCCGCUUUUCCAUCGCUUUUCCUCCAACCCUGCGGAAGUGCUCGCUG------UUGCGGGGUGAAAAACAAAAAUAAAUGCACCCAGCAGCAGCGGCAAUUUUAUUGGGUUU .................................(((((.(..(((.(((.(((((------((((.(((((.................))))).)))))))))))).)))..).))))). ( -37.63) >consensus GUUUUCGCCCCGCUUUUCCAUCGCUUUUCCUCCAACCCUGCGGAAGUGCUCGCUG______UUGCGGGGUGAAAAACAAAAAUAAAUGCACCCAGCAGCAGCGGCAAUUUUAUUGGGUUU .((((((((((((.......((((...............)))).(((....))).........))))))))))))((...(((((((((.....((....)).)))..))))))..)).. (-30.77 = -30.33 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:05 2006