
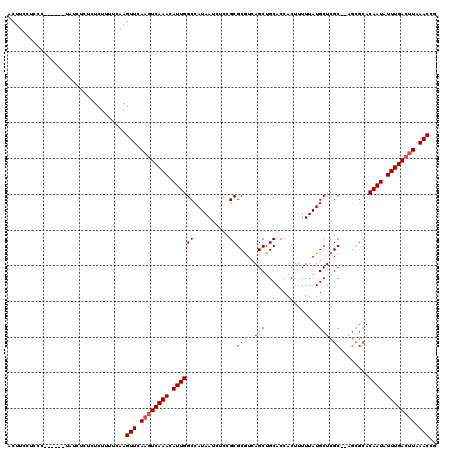
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,828,125 – 14,828,244 |
| Length | 119 |
| Max. P | 0.919702 |

| Location | 14,828,125 – 14,828,244 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.14 |
| Mean single sequence MFE | -19.07 |
| Consensus MFE | -14.16 |
| Energy contribution | -15.16 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14828125 119 + 27905053 ACUUCCUCCCCCUCUCUCUCUCUCUCUUUUCAAGUUCAAGUCAAACAUUGGCCAUAAUCUCGGCGCAUCAGCUGCACCACUUUUUAUGCUCGCCUAGCGCACAAUAUUUGAA-UAAACCG .................................(((....(((((.((((((.........((.(((.....))).)).........(((.....))))).)))).))))).-..))).. ( -16.70) >DroSec_CAF1 6286 107 + 1 ACUUCCUCCC------UAUCUCACUCUUUUCAAGUUCAAGUCAAACAUUGGCCAUAAUCUCCGCGCGUCAGCUGCACCACUUUUUAUGCUCGC-------ACAAUAUUUGACUUAAACCA ..........------.................(((.((((((((.(((((((((((.....((((....)).))........)))))...))-------.)))).)))))))).))).. ( -19.12) >DroSim_CAF1 6436 114 + 1 ACUUCCUCCC------UAUCUCUCUCUUUUCAAGUUCAAGUCAAACAUUGGCCAUAAUCUCCGCGCGUCAGCUGCACCACUUUUUAUGCUCGCACAGCGCACAAUAUUUGACUUAAACCG ..........------.................(((.((((((((.((((((..........((((....)).))............(((.....))))).)))).)))))))).))).. ( -21.40) >consensus ACUUCCUCCC______UAUCUCUCUCUUUUCAAGUUCAAGUCAAACAUUGGCCAUAAUCUCCGCGCGUCAGCUGCACCACUUUUUAUGCUCGC__AGCGCACAAUAUUUGACUUAAACCG .................................(((.((((((((.((((((..........))(((..(((...............))))))........)))).)))))))).))).. (-14.16 = -15.16 + 1.00)


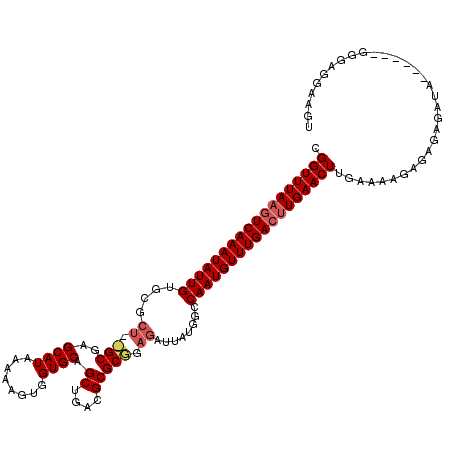
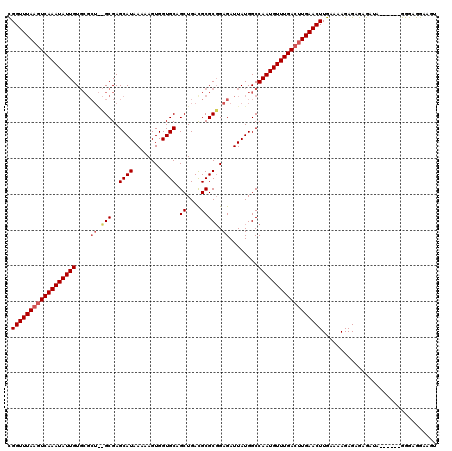
| Location | 14,828,125 – 14,828,244 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.14 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -25.99 |
| Energy contribution | -27.43 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14828125 119 - 27905053 CGGUUUA-UUCAAAUAUUGUGCGCUAGGCGAGCAUAAAAAGUGGUGCAGCUGAUGCGCCGAGAUUAUGGCCAAUGUUUGACUUGAACUUGAAAAGAGAGAGAGAGAGAGGGGGAGGAAGU .((((((-.((((((((((.((((...))...(((((....(((((((.....)))))))...)))))))))))))))))..))))))................................ ( -30.60) >DroSec_CAF1 6286 107 - 1 UGGUUUAAGUCAAAUAUUGU-------GCGAGCAUAAAAAGUGGUGCAGCUGACGCGCGGAGAUUAUGGCCAAUGUUUGACUUGAACUUGAAAAGAGUGAGAUA------GGGAGGAAGU .((((((((((((((((((.-------((...(((((....(.((((.......)))).)...)))))))))))))))))))))))))................------.......... ( -31.30) >DroSim_CAF1 6436 114 - 1 CGGUUUAAGUCAAAUAUUGUGCGCUGUGCGAGCAUAAAAAGUGGUGCAGCUGACGCGCGGAGAUUAUGGCCAAUGUUUGACUUGAACUUGAAAAGAGAGAGAUA------GGGAGGAAGU .((((((((((((((((((.((.(((((((.((((........))))......)))))))........))))))))))))))))))))................------.......... ( -34.40) >consensus CGGUUUAAGUCAAAUAUUGUGCGCU__GCGAGCAUAAAAAGUGGUGCAGCUGACGCGCGGAGAUUAUGGCCAAUGUUUGACUUGAACUUGAAAAGAGAGAGAUA______GGGAGGAAGU .((((((((((((((((((....((.(((..((((........)))).((....))))).))........))))))))))))))))))................................ (-25.99 = -27.43 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:03 2006