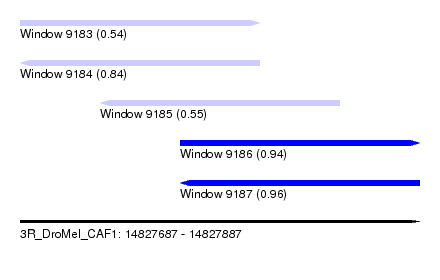
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,827,687 – 14,827,887 |
| Length | 200 |
| Max. P | 0.963843 |

| Location | 14,827,687 – 14,827,807 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.20 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -34.61 |
| Energy contribution | -34.61 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14827687 120 + 27905053 GACUCGACUUGAGACUCUUCCACGAAAUGGCGAAACCCGUUUUGGGUCUCAGCCUCUCGGGUCCAAGAGUGUCUUUCAAUUGUAUUCAAUGCUAACUCAAUUGCUCUGGCCGAGGGCUAC ..........(((((((......(((((((......))))))))))))))((((.(((((..(((.(((((..........((((...)))).........))))))))))))))))).. ( -36.01) >DroSec_CAF1 5853 117 + 1 ---UCGACUUGAGACUCUUCCACGAAAUGGCGAAACCCGUUUUGGGUCUCAGCCUCUCGGGUCCAAGAGUGUCUUUCAAUUGUAUUCAAUGCUAACUCAAUUGCUCUGGCCGAGGGCUAC ---.......(((((((......(((((((......))))))))))))))((((.(((((..(((.(((((..........((((...)))).........))))))))))))))))).. ( -36.01) >DroSim_CAF1 6004 117 + 1 ---UUGACUUGAGACUCUUCCACGAAAUGGCGAAACCCGUUUUGGGUCUCAGCCUCUCCGGUCCAAGAGUGUCUUUCAAUUGUAUUCAAUGCUAACUCAAUUGCUCUGGCCGAGGGCUAC ---.......(((((((......(((((((......))))))))))))))((((.(((.((.(((.(((((..........((((...)))).........))))))))))))))))).. ( -34.61) >consensus ___UCGACUUGAGACUCUUCCACGAAAUGGCGAAACCCGUUUUGGGUCUCAGCCUCUCGGGUCCAAGAGUGUCUUUCAAUUGUAUUCAAUGCUAACUCAAUUGCUCUGGCCGAGGGCUAC ..........(((((((......(((((((......))))))))))))))((((.(((.((.(((.(((((..........((((...)))).........))))))))))))))))).. (-34.61 = -34.61 + -0.00)



| Location | 14,827,687 – 14,827,807 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.20 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -36.00 |
| Energy contribution | -36.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14827687 120 - 27905053 GUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACAAUUGAAAGACACUCUUGGACCCGAGAGGCUGAGACCCAAAACGGGUUUCGCCAUUUCGUGGAAGAGUCUCAAGUCGAGUC ......((((((..(((((((((.(((.......))).))))))......((((((...((((((((((.(((((((.....)))))))))).))))).)))))))))))..)))))).. ( -43.30) >DroSec_CAF1 5853 117 - 1 GUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACAAUUGAAAGACACUCUUGGACCCGAGAGGCUGAGACCCAAAACGGGUUUCGCCAUUUCGUGGAAGAGUCUCAAGUCGA--- .......(((((..(((((((((.(((.......))).))))))......((((((...((((((((((.(((((((.....)))))))))).))))).)))))))))))..)))))--- ( -39.90) >DroSim_CAF1 6004 117 - 1 GUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACAAUUGAAAGACACUCUUGGACCGGAGAGGCUGAGACCCAAAACGGGUUUCGCCAUUUCGUGGAAGAGUCUCAAGUCAA--- .........(((..(((((((((.(((.......))).))))))......((((((...((((((((((.(((((((.....)))))))))).)))).))))))))))))..)))..--- ( -35.30) >consensus GUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACAAUUGAAAGACACUCUUGGACCCGAGAGGCUGAGACCCAAAACGGGUUUCGCCAUUUCGUGGAAGAGUCUCAAGUCGA___ .......(((((..(((((((((.(((.......))).))))))......((((((...((((((((((.(((((((.....)))))))))).))))).)))))))))))..)))))... (-36.00 = -36.67 + 0.67)

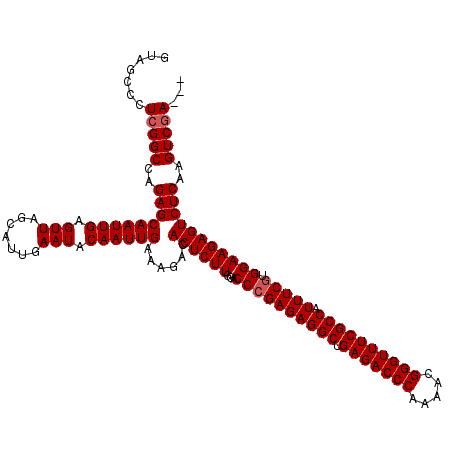

| Location | 14,827,727 – 14,827,847 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -29.74 |
| Energy contribution | -30.08 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14827727 120 - 27905053 UCGGUUAGACAGCCAGCCAAAGCUAUGGCCAUAUAACCAUGUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACAAUUGAAAGACACUCUUGGACCCGAGAGGCUGAGACCCAAA ..(((((....((((((....))..))))....)))))...((((((((((((((((((((((.(((.......))).)))))).......))).)))..))))).)))))......... ( -35.91) >DroSec_CAF1 5890 116 - 1 UCGGUUAGACAG----CCAAAGCUAUGGCCAUAUAACCAUGUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACAAUUGAAAGACACUCUUGGACCCGAGAGGCUGAGACCCAAA ..(((((....(----(((......))))....)))))...((((((((((((((((((((((.(((.......))).)))))).......))).)))..))))).)))))......... ( -35.31) >DroSim_CAF1 6041 120 - 1 UCGGUUAGACAGCCAGCCAAAGCUAUGGCCAUAUAACCAUGUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACAAUUGAAAGACACUCUUGGACCGGAGAGGCUGAGACCCAAA ..(((((....((((((....))..))))....)))))...((((((((((((((((((((((.(((.......))).)))))).......))).))).)).))).)))))......... ( -34.51) >consensus UCGGUUAGACAGCCAGCCAAAGCUAUGGCCAUAUAACCAUGUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACAAUUGAAAGACACUCUUGGACCCGAGAGGCUGAGACCCAAA ..(((((........((((......))))....)))))...((((((((((((((((((((((.(((.......))).)))))).......))).))).)).))).)))))......... (-29.74 = -30.08 + 0.33)

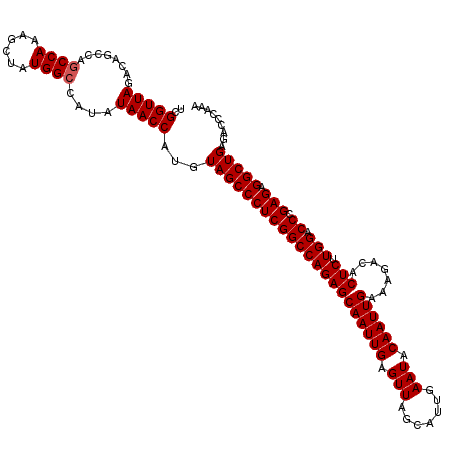

| Location | 14,827,767 – 14,827,887 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -49.93 |
| Consensus MFE | -44.84 |
| Energy contribution | -45.73 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -4.65 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14827767 120 + 27905053 UGUAUUCAAUGCUAACUCAAUUGCUCUGGCCGAGGGCUACAUGGUUAUAUGGCCAUAGCUUUGGCUGGCUGUCUAACCGAUUCCAUUUCGCUUUGGCAGGGGCAAUUGAGGCACACUCUA .........(((...((((((((((((.(((((((.......(((((.(((((((..((....))))))))).)))))((.......)).))))))).)))))))))))))))....... ( -46.80) >DroSec_CAF1 5930 116 + 1 UGUAUUCAAUGCUAACUCAAUUGCUCUGGCCGAGGGCUACAUGGUUAUAUGGCCAUAGCUUUGG----CUGUCUAACCGAUUCCAUUUCGCCCUGGCAGGGGCAAUUGAGGCACACUCGA .........(((...((((((((((((.(((.(((((.....(((((.(((((((......)))----)))).)))))((.......)))))))))).)))))))))))))))....... ( -51.20) >DroSim_CAF1 6081 120 + 1 UGUAUUCAAUGCUAACUCAAUUGCUCUGGCCGAGGGCUACAUGGUUAUAUGGCCAUAGCUUUGGCUGGCUGUCUAACCGAUUCCAUUUCGCCCUGGCAGGGGCAAUUGAGGCACACUCGA .........(((...((((((((((((.(((.(((((.....(((((.(((((((..((....))))))))).)))))((.......)))))))))).)))))))))))))))....... ( -51.80) >consensus UGUAUUCAAUGCUAACUCAAUUGCUCUGGCCGAGGGCUACAUGGUUAUAUGGCCAUAGCUUUGGCUGGCUGUCUAACCGAUUCCAUUUCGCCCUGGCAGGGGCAAUUGAGGCACACUCGA .........(((...((((((((((((.(((.(((((.....(((((.(((((((..((....))))))))).)))))((.......)))))))))).)))))))))))))))....... (-44.84 = -45.73 + 0.89)

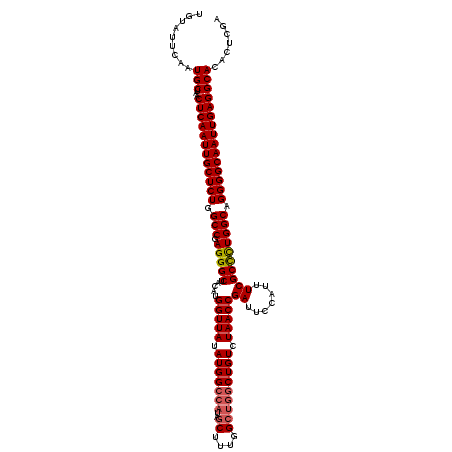
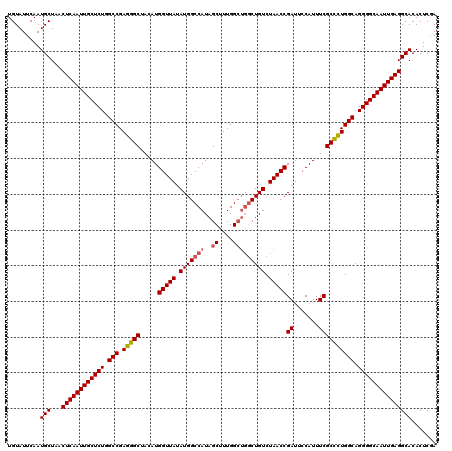
| Location | 14,827,767 – 14,827,887 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -38.43 |
| Energy contribution | -39.43 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14827767 120 - 27905053 UAGAGUGUGCCUCAAUUGCCCCUGCCAAAGCGAAAUGGAAUCGGUUAGACAGCCAGCCAAAGCUAUGGCCAUAUAACCAUGUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACA ...(((((..(((((((((..(((((...((...((((....((((....)))).((((......)))).......))))...))....)).))).)))))))))...)))))....... ( -37.20) >DroSec_CAF1 5930 116 - 1 UCGAGUGUGCCUCAAUUGCCCCUGCCAGGGCGAAAUGGAAUCGGUUAGACAG----CCAAAGCUAUGGCCAUAUAACCAUGUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACA ((.(((((..(((((((((..((((((((((...((((....(((((...((----(....))).)))))......))))...))))).)).))).)))))))))...)))))))..... ( -44.20) >DroSim_CAF1 6081 120 - 1 UCGAGUGUGCCUCAAUUGCCCCUGCCAGGGCGAAAUGGAAUCGGUUAGACAGCCAGCCAAAGCUAUGGCCAUAUAACCAUGUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACA ((.(((((..(((((((((..((((((((((...((((....((((....)))).((((......)))).......))))...))))).)).))).)))))))))...)))))))..... ( -47.30) >consensus UCGAGUGUGCCUCAAUUGCCCCUGCCAGGGCGAAAUGGAAUCGGUUAGACAGCCAGCCAAAGCUAUGGCCAUAUAACCAUGUAGCCCUCGGCCAGAGCAAUUGAGUUAGCAUUGAAUACA ...(((((..(((((((((..((((((((((...((((....(((((...(((........))).)))))......))))...))))).)).))).)))))))))...)))))....... (-38.43 = -39.43 + 1.00)


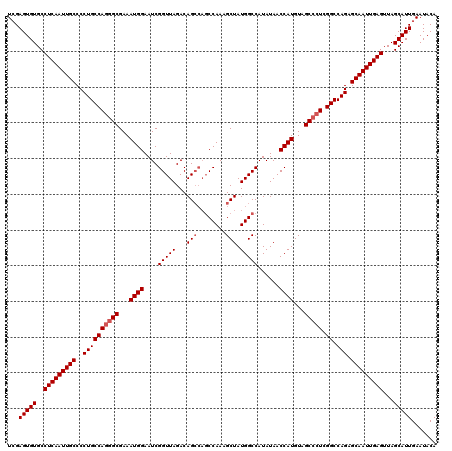
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:02 2006