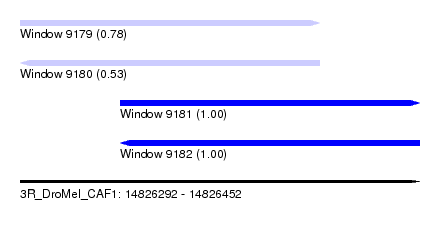
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,826,292 – 14,826,452 |
| Length | 160 |
| Max. P | 0.999999 |

| Location | 14,826,292 – 14,826,412 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -33.50 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14826292 120 + 27905053 UCCGCACCUUUGGCUGGGUGGCUGUUCAGCGGGCAAAGUGUUGCCAAAUGUUUGCAGCAUUUCCUGUUGUCUACGGGGCGUUGUCGCUGCAAAGUUUUGGUCUUUUAACUUUUAGCACGU .((.(((((......)))))(((....)))))(((((((...((((((..((((((((((.((((((.....))))))....)).))))))))..))))))......)))))..)).... ( -39.20) >DroSec_CAF1 4476 116 + 1 UCCGCACCUUUGGCU----GGCUGUUCAGCGGGCAAAGUGUUGCCAAAUGUUUGCAGCAUUUCCUGUUGUCUACGGGGCGUUGUCGCUGCAAAGUUUUGGUCUUUUAACUUUUAGCACGU ...(((((((((.((----.(((....))).))))))).)).((((((..((((((((((.((((((.....))))))....)).))))))))..)))))).............)).... ( -37.90) >DroSim_CAF1 4592 116 + 1 UCCGCACCUUUGGCU----GGCUGUUCAGCGGGCAAAGUGUUGCCAAAUGUUUGCAGCAUUUCCUGUUGUCUACGGGGCGUUGUCGCUGCAAAGUUUUGGUCNNNNNNNNNNNNNNNNNN ...((((.((((.((----.(((....))).)))))))))).((((((..((((((((((.((((((.....))))))....)).))))))))..))))))................... ( -36.50) >consensus UCCGCACCUUUGGCU____GGCUGUUCAGCGGGCAAAGUGUUGCCAAAUGUUUGCAGCAUUUCCUGUUGUCUACGGGGCGUUGUCGCUGCAAAGUUUUGGUCUUUUAACUUUUAGCACGU ...((((.((((.((.....(((....))))).)))))))).((((((..((((((((((.((((((.....))))))....)).))))))))..))))))................... (-33.50 = -33.50 + 0.00)

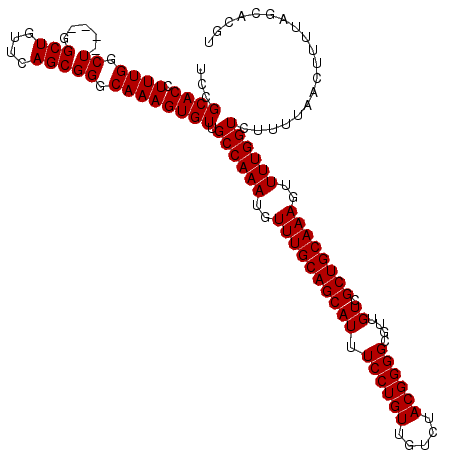
| Location | 14,826,292 – 14,826,412 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -23.95 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14826292 120 - 27905053 ACGUGCUAAAAGUUAAAAGACCAAAACUUUGCAGCGACAACGCCCCGUAGACAACAGGAAAUGCUGCAAACAUUUGGCAACACUUUGCCCGCUGAACAGCCACCCAGCCAAAGGUGCGGA .((..(............(.(((((..(((((((((......((..((.....)).))...)))))))))..))))))....(((((...((((..........))))))))))..)).. ( -30.00) >DroSec_CAF1 4476 116 - 1 ACGUGCUAAAAGUUAAAAGACCAAAACUUUGCAGCGACAACGCCCCGUAGACAACAGGAAAUGCUGCAAACAUUUGGCAACACUUUGCCCGCUGAACAGCC----AGCCAAAGGUGCGGA .((..(............(.(((((..(((((((((......((..((.....)).))...)))))))))..))))))....(((((...((((......)----)))))))))..)).. ( -31.10) >DroSim_CAF1 4592 116 - 1 NNNNNNNNNNNNNNNNNNGACCAAAACUUUGCAGCGACAACGCCCCGUAGACAACAGGAAAUGCUGCAAACAUUUGGCAACACUUUGCCCGCUGAACAGCC----AGCCAAAGGUGCGGA ....................(((((..(((((((((......((..((.....)).))...)))))))))..)))))(..((((((....((((......)----)))..))))))..). ( -27.50) >consensus ACGUGCUAAAAGUUAAAAGACCAAAACUUUGCAGCGACAACGCCCCGUAGACAACAGGAAAUGCUGCAAACAUUUGGCAACACUUUGCCCGCUGAACAGCC____AGCCAAAGGUGCGGA ..................(.(((((..(((((((((......((..((.....)).))...)))))))))..))))))..........((((......((......)).......)))). (-23.95 = -23.95 + 0.00)


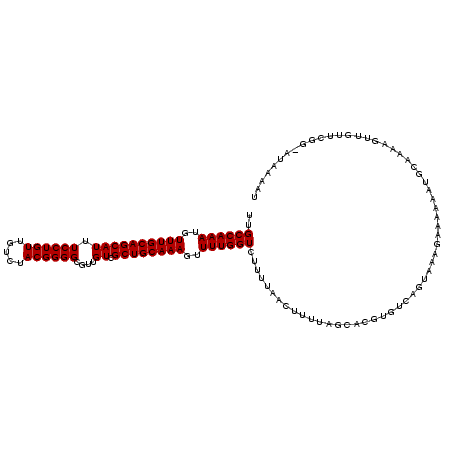
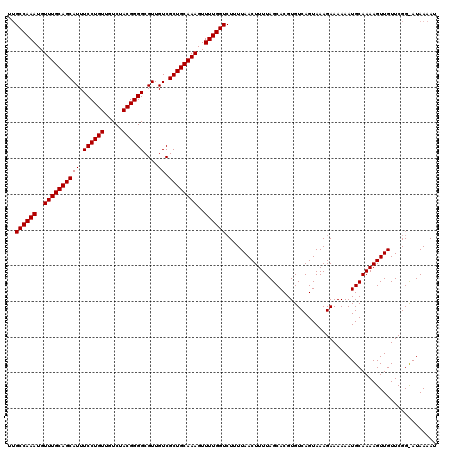
| Location | 14,826,332 – 14,826,452 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.50 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 5.69 |
| SVM RNA-class probability | 0.999992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14826332 120 + 27905053 UUGCCAAAUGUUUGCAGCAUUUCCUGUUGUCUACGGGGCGUUGUCGCUGCAAAGUUUUGGUCUUUUAACUUUUAGCACGUGUCAGUAAAGAAAAAAUGCAAAAGUUGUUCGGUAUAAAAU ..((((((..((((((((((.((((((.....))))))....)).))))))))..))))))....((((((((.(((....((......)).....)))))))))))............. ( -32.10) >DroSec_CAF1 4512 120 + 1 UUGCCAAAUGUUUGCAGCAUUUCCUGUUGUCUACGGGGCGUUGUCGCUGCAAAGUUUUGGUCUUUUAACUUUUAGCACGUGUCAGUAAAGAAAAAAUGCAAAAGUUGUUCGGCAUAAAAU ..((((((..((((((((((.((((((.....))))))....)).))))))))..))))))....((((((((.(((....((......)).....)))))))))))............. ( -32.10) >DroSim_CAF1 4628 120 + 1 UUGCCAAAUGUUUGCAGCAUUUCCUGUUGUCUACGGGGCGUUGUCGCUGCAAAGUUUUGGUCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ..((((((..((((((((((.((((((.....))))))....)).))))))))..))))))........................................................... ( -23.80) >consensus UUGCCAAAUGUUUGCAGCAUUUCCUGUUGUCUACGGGGCGUUGUCGCUGCAAAGUUUUGGUCUUUUAACUUUUAGCACGUGUCAGUAAAGAAAAAAUGCAAAAGUUGUUCGG_AUAAAAU ..((((((..((((((((((.((((((.....))))))....)).))))))))..))))))........................................................... (-23.80 = -23.80 + 0.00)

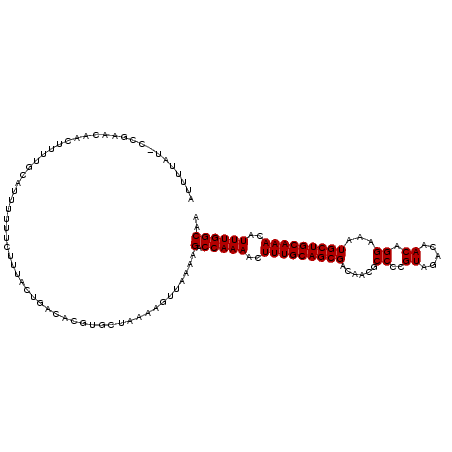
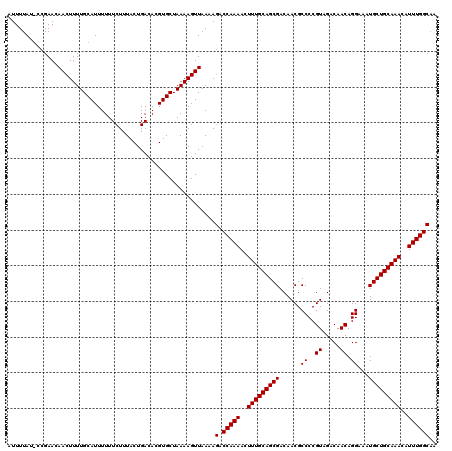
| Location | 14,826,332 – 14,826,452 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.50 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.72 |
| SVM decision value | 6.55 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14826332 120 - 27905053 AUUUUAUACCGAACAACUUUUGCAUUUUUUCUUUACUGACACGUGCUAAAAGUUAAAAGACCAAAACUUUGCAGCGACAACGCCCCGUAGACAACAGGAAAUGCUGCAAACAUUUGGCAA ..............(((((((((((....((......))...)))).)))))))....(.(((((..(((((((((......((..((.....)).))...)))))))))..)))))).. ( -25.60) >DroSec_CAF1 4512 120 - 1 AUUUUAUGCCGAACAACUUUUGCAUUUUUUCUUUACUGACACGUGCUAAAAGUUAAAAGACCAAAACUUUGCAGCGACAACGCCCCGUAGACAACAGGAAAUGCUGCAAACAUUUGGCAA ......(((((((.(((((((((((....((......))...)))).))))))).............(((((((((......((..((.....)).))...)))))))))..))))))). ( -28.80) >DroSim_CAF1 4628 120 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGACCAAAACUUUGCAGCGACAACGCCCCGUAGACAACAGGAAAUGCUGCAAACAUUUGGCAA ..........................................................(.(((((..(((((((((......((..((.....)).))...)))))))))..)))))).. ( -16.90) >consensus AUUUUAU_CCGAACAACUUUUGCAUUUUUUCUUUACUGACACGUGCUAAAAGUUAAAAGACCAAAACUUUGCAGCGACAACGCCCCGUAGACAACAGGAAAUGCUGCAAACAUUUGGCAA ..........................................................(.(((((..(((((((((......((..((.....)).))...)))))))))..)))))).. (-17.03 = -17.03 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:57 2006