
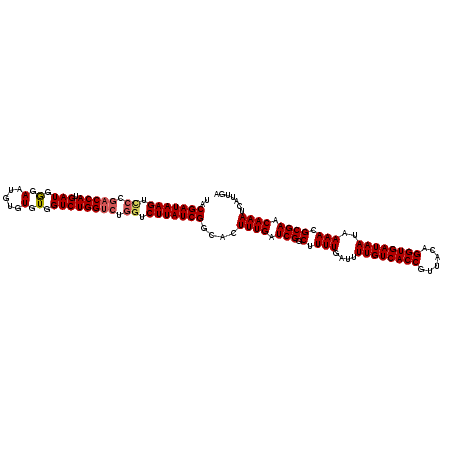
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,823,299 – 14,823,415 |
| Length | 116 |
| Max. P | 0.824149 |

| Location | 14,823,299 – 14,823,415 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -37.11 |
| Consensus MFE | -32.86 |
| Energy contribution | -33.87 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14823299 116 + 27905053 UACGAUAAGUCCCGACCAUGAUGAGAAUGUGUGUGGUCUGGC----UCUUAUCGGCACUUUGAUCGGCUUUUGAUUUUGUCACCGUUACAGGUGAUAAUAAAACGCGAACAAAUCAUUGA ..(((((((..((((((((.............)))))).)).----.)))))))....((((.(((.(.(((....((((((((......))))))))..))).)))).))))....... ( -34.62) >DroSec_CAF1 1439 120 + 1 UCCGAUAAGUUCCGACCAUGAUGGGAAUGUGUGUGGUCUGGUCUGGUCUUAUCGGCACUUUGAUCGGCUUUUGAUUUUGUCACCGUUACAGGUGAUAAUAAAACGCGAACAAAUCAUUGA .((((((((..(((((((.(((.(.(.....).).)))))))).)).))))))))...((((.(((.(.(((....((((((((......))))))))..))).)))).))))....... ( -37.90) >DroSim_CAF1 1577 120 + 1 UACGAUAAGUCCCGACCAUGAUGGGAAUGUGUGUGGUCUGGUCUGGUCUUAUCGGCACUUUGAUCGGCUUUUGAUUUUGUCACCGUUACAGGUGAUAAUAAAACGCGAACAAAUCAUUGA ..(((((((.((.(((((.(((.(.(.....).).)))))))).)).)))))))....((((.(((.(.(((....((((((((......))))))))..))).)))).))))....... ( -38.80) >consensus UACGAUAAGUCCCGACCAUGAUGGGAAUGUGUGUGGUCUGGUCUGGUCUUAUCGGCACUUUGAUCGGCUUUUGAUUUUGUCACCGUUACAGGUGAUAAUAAAACGCGAACAAAUCAUUGA ..(((((((.((.(((((.(((.(.(.....).).)))))))).)).)))))))....((((.(((.(.(((....((((((((......))))))))..))).)))).))))....... (-32.86 = -33.87 + 1.00)

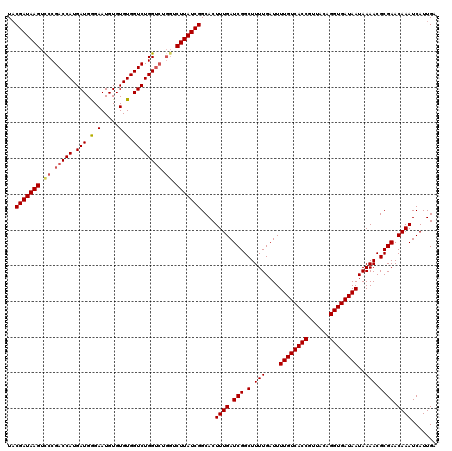
| Location | 14,823,299 – 14,823,415 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -25.31 |
| Energy contribution | -25.43 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14823299 116 - 27905053 UCAAUGAUUUGUUCGCGUUUUAUUAUCACCUGUAACGGUGACAAAAUCAAAAGCCGAUCAAAGUGCCGAUAAGA----GCCAGACCACACACAUUCUCAUCAUGGUCGGGACUUAUCGUA .......((((.(((.((((((((.(((((......)))))...)))..)))))))).))))....(((((((.----.((.(((((...............))))).)).))))))).. ( -28.36) >DroSec_CAF1 1439 120 - 1 UCAAUGAUUUGUUCGCGUUUUAUUAUCACCUGUAACGGUGACAAAAUCAAAAGCCGAUCAAAGUGCCGAUAAGACCAGACCAGACCACACACAUUCCCAUCAUGGUCGGAACUUAUCGGA .......((((.(((.((((((((.(((((......)))))...)))..)))))))).))))...((((((((.((.(((((((...............)).)))))))..)))))))). ( -30.96) >DroSim_CAF1 1577 120 - 1 UCAAUGAUUUGUUCGCGUUUUAUUAUCACCUGUAACGGUGACAAAAUCAAAAGCCGAUCAAAGUGCCGAUAAGACCAGACCAGACCACACACAUUCCCAUCAUGGUCGGGACUUAUCGUA .......((((.(((.((((((((.(((((......)))))...)))..)))))))).))))....(((((((.((.(((((((...............)).))))).)).))))))).. ( -29.86) >consensus UCAAUGAUUUGUUCGCGUUUUAUUAUCACCUGUAACGGUGACAAAAUCAAAAGCCGAUCAAAGUGCCGAUAAGACCAGACCAGACCACACACAUUCCCAUCAUGGUCGGGACUUAUCGUA .......((((.(((.((((((((.(((((......)))))...)))..)))))))).))))....(((((((....(((((((...............)).)))))....))))))).. (-25.31 = -25.43 + 0.11)


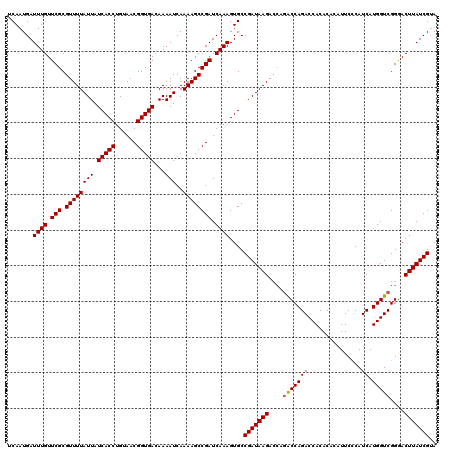
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:46 2006