
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,814,768 – 14,814,940 |
| Length | 172 |
| Max. P | 0.914602 |

| Location | 14,814,768 – 14,814,866 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.76 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -20.50 |
| Energy contribution | -23.12 |
| Covariance contribution | 2.62 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14814768 98 - 27905053 GCAACGCAACCCGUUGCUGCCGUCGAUAUUGGCGAU--------AUGGCUGCUCUUGCCAAUCUCGAGAAGGCACGCAACAAGCAAUUCCAGGCAAGAAUCUCCUC ............(((((((((.((((.((((((((.--------..........)))))))).))))...)))).)))))..((........))............ ( -29.20) >DroSec_CAF1 7375 106 - 1 GCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAUAUUGGGAUAUGGCUGCUGUUGCCAAUCUCGAGUAGGCACGCAACAAGCCAUUCCAGGCAAGAAUCUGCAC .....(((....(((((((((((((....))))....(((((((.((((.......)))))))))))...)))).)))))..(((......))).......))).. ( -40.20) >DroSim_CAF1 7405 106 - 1 GCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAUAUUGGGAUAUGGCUGCUGUUGCCAAUCUCGAGUAGGCACGCAACAAGCCAUUCCAGGCAAGAAUCUCCAC ((((((.....))))))((((((((....)))).......(((.((((((..(((((((...........)))).)))...))))))))).))))........... ( -37.60) >DroEre_CAF1 9132 87 - 1 GCAACGCAACCCGUUGCUGCCGCCGAUAUU-----------------GCUGCUGUUGCCAU--UCGAGGAAGCACGCAACAAGCAAUUUCAGGGAAGAAUCUCCUU ((((((.....)))))).......((.(((-----------------(((..((((((..(--((...)))....)))))))))))).))(((((......))))) ( -25.00) >consensus GCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAU________AUGGCUGCUGUUGCCAAUCUCGAGUAGGCACGCAACAAGCAAUUCCAGGCAAGAAUCUCCAC ((((((.....))))))((((((((....))))...........((((((..((((((...(((.....)))...))))))))))))....))))........... (-20.50 = -23.12 + 2.62)



| Location | 14,814,796 – 14,814,904 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.65 |
| Mean single sequence MFE | -40.75 |
| Consensus MFE | -35.66 |
| Energy contribution | -37.73 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14814796 108 + 27905053 UGCGUGCCUUCUCGAGAUUGGCAAGAGCAGCCAU--------AUCGCCAAUAUCGACGGCAGCAACGGGUUGCGUUGCUGUUGCGUCUGGAAUCUGCAGAUUGAUUGCCGAUCGCU (((.((((.((....))..))))...)))((...--------((((.((((..((.(((((((((((.....)))))))))))))((((.......))))...)))).)))).)). ( -37.40) >DroSec_CAF1 7403 116 + 1 UGCGUGCCUACUCGAGAUUGGCAACAGCAGCCAUAUCCCAAUAUCGCCAAUAUCGGCGGCAGCAACGGGUUGCGUUGCUGUUGCGUCUGGAAUCUGCAGAUUGAUUGCCGAUCGCU .((.(((....((.((((..(((((((((((.............((((......))))(((((.....)))))))))))))))))))).))....)))(((((.....))))))). ( -45.60) >DroSim_CAF1 7433 116 + 1 UGCGUGCCUACUCGAGAUUGGCAACAGCAGCCAUAUCCCAAUAUCGCCAAUAUCGGCGGCAGCAACGGGUUGCGUUGCUGUUGCGUCUGGAAUCUGCAGAUUGAUUGCCGAUCGCU .((.(((....((.((((..(((((((((((.............((((......))))(((((.....)))))))))))))))))))).))....)))(((((.....))))))). ( -45.60) >DroEre_CAF1 9160 97 + 1 UGCGUGCUUCCUCGA--AUGGCAACAGCAGC-----------------AAUAUCGGCGGCAGCAACGGGUUGCGUUGCUGUUGCGUCUGGAAUCUGUAGAUUGAUUGCCGAUCGCU (((.((((.......--..))))...)))((-----------------.......((((((((((((.....))))))))))))(((.(.((((........)))).).))).)). ( -34.40) >consensus UGCGUGCCUACUCGAGAUUGGCAACAGCAGCCAU________AUCGCCAAUAUCGGCGGCAGCAACGGGUUGCGUUGCUGUUGCGUCUGGAAUCUGCAGAUUGAUUGCCGAUCGCU .((.(((....((.((((..(((((((((((.............((((......))))(((((.....)))))))))))))))))))).))....)))(((((.....))))))). (-35.66 = -37.73 + 2.06)



| Location | 14,814,796 – 14,814,904 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.65 |
| Mean single sequence MFE | -38.85 |
| Consensus MFE | -30.14 |
| Energy contribution | -30.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14814796 108 - 27905053 AGCGAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGUCGAUAUUGGCGAU--------AUGGCUGCUCUUGCCAAUCUCGAGAAGGCACGCA .(((.((((.((((........)))))).)))))...(((((((.....)))))))(((.((((.((((((((.--------..........)))))))).))))...)))..... ( -36.80) >DroSec_CAF1 7403 116 - 1 AGCGAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAUAUUGGGAUAUGGCUGCUGUUGCCAAUCUCGAGUAGGCACGCA .(((((((((......((((.......)))).....((((((((.....))))))))..))))))....((....(((((((.((((.......)))))))))))....)).))). ( -43.20) >DroSim_CAF1 7433 116 - 1 AGCGAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAUAUUGGGAUAUGGCUGCUGUUGCCAAUCUCGAGUAGGCACGCA .(((((((((......((((.......)))).....((((((((.....))))))))..))))))....((....(((((((.((((.......)))))))))))....)).))). ( -43.20) >DroEre_CAF1 9160 97 - 1 AGCGAUCGGCAAUCAAUCUACAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUU-----------------GCUGCUGUUGCCAU--UCGAGGAAGCACGCA .(((((((((....((((....))))..........((((((((.....))))))))..))))))...-----------------..((((....((..--....)).))))))). ( -32.20) >consensus AGCGAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAU________AUGGCUGCUGUUGCCAAUCUCGAGUAGGCACGCA .(((((((((......((((.......)))).....((((((((.....))))))))..))))))..................((((.......))))..............))). (-30.14 = -30.70 + 0.56)

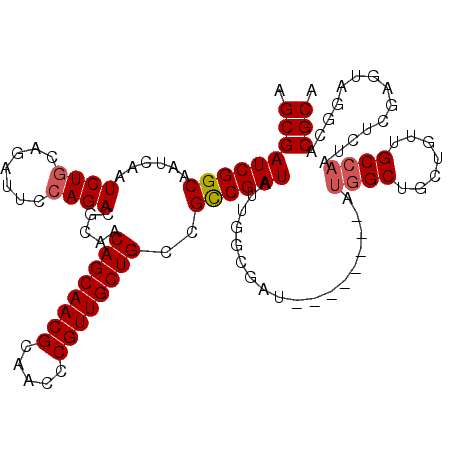

| Location | 14,814,830 – 14,814,940 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 87.61 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -28.23 |
| Energy contribution | -30.04 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14814830 110 - 27905053 UAGGCGAACGUGCAAAAAUGCGGCAUGAGCGGCCUAAGCGAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGUCGAUAUUGGCGAU---- ((((((..(((((.........)))))..).))))).(((.((((.((((........)))))).)))))..((((((((.....)))))))).(((((....)))))..---- ( -36.60) >DroSec_CAF1 7441 114 - 1 UAGGCGAACGUGCAAAAAUGCGGCAUGAGCGGCCUAAGCGAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAUAUUG ((((((..(((((.........)))))..).))))).(((.((((.((((........)))))).)))))..((((((((.....)))))))).(((((....)))))...... ( -39.30) >DroSim_CAF1 7471 114 - 1 UAGGCGAACGUGCAAAAAUGCGGCAUGAGCGGCCUAAGCGAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAUAUUG ((((((..(((((.........)))))..).))))).(((.((((.((((........)))))).)))))..((((((((.....)))))))).(((((....)))))...... ( -39.30) >DroEre_CAF1 9189 104 - 1 UAGGCGAACGUGCAAAAAUGCGGCAUGAGCGGUCUAAGCGAUCGGCAAUCAAUCUACAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUU---------- ..((((..(((((.........))))).((((((.....))))((.((((........))))))....))..((((((((.....)))))))).))))......---------- ( -33.50) >DroYak_CAF1 8216 89 - 1 UAGGCGAACGUGCAAAAAUGCGGCAUAAGCGGCCUAAGCGAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAAAAGCAACGCAACCCGUU------------------------- ...(((....(((......((.((....)).))....(((.((((.((((........)))))).)))))....))).)))........------------------------- ( -21.60) >consensus UAGGCGAACGUGCAAAAAUGCGGCAUGAGCGGCCUAAGCGAUCGGCAAUCAAUCUGCAGAUUCCAGACGCAACAGCAACGCAACCCGUUGCUGCCGCCGAUAUUGGCGAU____ ((((((..(((((.........)))))..).))))).(((.((((.((((........)))))).)))))..((((((((.....)))))))).(((((....)))))...... (-28.23 = -30.04 + 1.81)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:34 2006