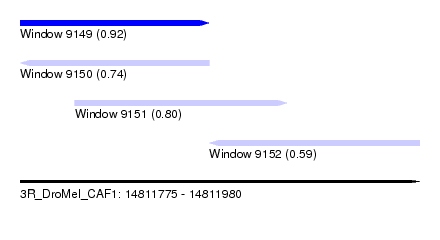
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,811,775 – 14,811,980 |
| Length | 205 |
| Max. P | 0.918797 |

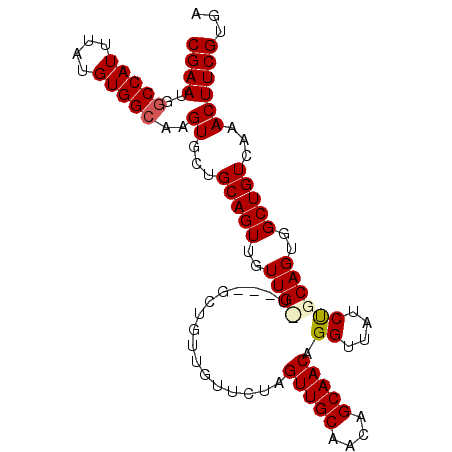
| Location | 14,811,775 – 14,811,872 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.06 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14811775 97 + 27905053 CGAAUGGCCAUUUAUGUGGCAAGUGCUGCAGUUGUUGUU---GCUGUUGUUCUAGUUGCAACAGCAACAGGUUAUCCGCAGUGGCUGUCAAACUUCGUGA ((((..(((((....)))))..((...(((((..(((((---((((((((.......))))))))))).((....))..))..)))))...))))))... ( -36.70) >DroSec_CAF1 4389 100 + 1 CGAAUGUCCAUUUAUGUGGCAAGUGCUGCAGUUGUUGCUGUAGCUGUUGUUCUAGUUGCAACAGCAACAGGUUAUCUGCAGUGGCUGUCAAACUUCGUGA ........(((.....(((((..((((((((.((...((((.((((((((.......)))))))).))))..)).))))))))..)))))......))). ( -35.00) >DroSim_CAF1 4397 100 + 1 CGAAUGGCCAUUUAUGUGGCAAGUGCUGCAGUUGUUGUUGUAGCUGUUGUUCUAGUUGCAACAGCAACAGGUUAUCUGCAGUGGCUGUCAAACUUCGUGA .((.((((((((..((..((....))..))((((((((((((((((......))))))))))))))))...........))))))))))........... ( -38.70) >DroEre_CAF1 6153 91 + 1 CGAAUGGCCAUUUAUGUGGCAAGUGCUGCAGUUGUUG---------UUGUUCUAGUUGCAACAGCAACAGGUUAUCUGCAGUGGCUGUCAAACUUCGUGA .((.((((((((..((..((....))..))(((((((---------((((.......)))))))))))...........))))))))))........... ( -31.50) >DroYak_CAF1 4623 91 + 1 CGAAUGGCCAUUUAUGUGGCAAGUGCCGCAGUUGUUG---------UUGUUCUAGUUGCAACAGCAACAGGUUAUCUGCAGUGGCUGUCAAACUUCGUGA .((.((((((((..((((((....))))))(((((((---------((((.......)))))))))))...........))))))))))........... ( -34.30) >consensus CGAAUGGCCAUUUAUGUGGCAAGUGCUGCAGUUGUUG_U___GCUGUUGUUCUAGUUGCAACAGCAACAGGUUAUCUGCAGUGGCUGUCAAACUUCGUGA ((((..(((((....)))))..((...(((((..((((................(((((....))))).((....))))))..)))))...))))))... (-25.70 = -26.06 + 0.36)

| Location | 14,811,775 – 14,811,872 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -18.22 |
| Energy contribution | -18.10 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14811775 97 - 27905053 UCACGAAGUUUGACAGCCACUGCGGAUAACCUGUUGCUGUUGCAACUAGAACAACAGC---AACAACAACUGCAGCACUUGCCACAUAAAUGGCCAUUCG ...((((............((((((......(((((((((((.........)))))))---))))....)))))).....((((......))))..)))) ( -29.10) >DroSec_CAF1 4389 100 - 1 UCACGAAGUUUGACAGCCACUGCAGAUAACCUGUUGCUGUUGCAACUAGAACAACAGCUACAGCAACAACUGCAGCACUUGCCACAUAAAUGGACAUUCG ...((((............((((((.....((((.(((((((.........))))))).))))......))))))....(((((......))).)))))) ( -26.70) >DroSim_CAF1 4397 100 - 1 UCACGAAGUUUGACAGCCACUGCAGAUAACCUGUUGCUGUUGCAACUAGAACAACAGCUACAACAACAACUGCAGCACUUGCCACAUAAAUGGCCAUUCG ...((((............((((((......(((.(((((((.........))))))).))).......)))))).....((((......))))..)))) ( -25.92) >DroEre_CAF1 6153 91 - 1 UCACGAAGUUUGACAGCCACUGCAGAUAACCUGUUGCUGUUGCAACUAGAACAA---------CAACAACUGCAGCACUUGCCACAUAAAUGGCCAUUCG ...((((............((((((......(((((.((((........)))).---------))))).)))))).....((((......))))..)))) ( -20.30) >DroYak_CAF1 4623 91 - 1 UCACGAAGUUUGACAGCCACUGCAGAUAACCUGUUGCUGUUGCAACUAGAACAA---------CAACAACUGCGGCACUUGCCACAUAAAUGGCCAUUCG ...(((((.....).((((..((((......(((((.((((........)))).---------))))).))))(((....))).......))))..)))) ( -21.20) >consensus UCACGAAGUUUGACAGCCACUGCAGAUAACCUGUUGCUGUUGCAACUAGAACAACAGC___A_CAACAACUGCAGCACUUGCCACAUAAAUGGCCAUUCG ...(((((((((.(((...))))))))....((((((.((((........................)))).))))))...((((......))))..)))) (-18.22 = -18.10 + -0.12)

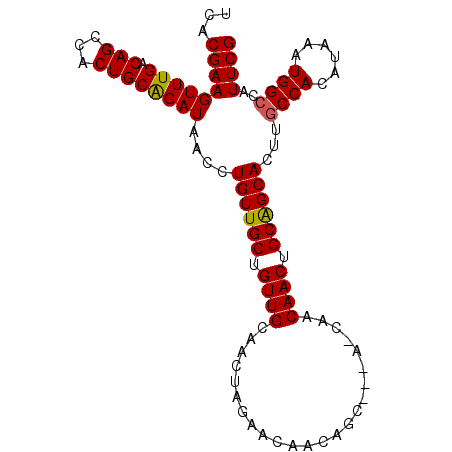
| Location | 14,811,803 – 14,811,912 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.87 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -21.80 |
| Energy contribution | -25.80 |
| Covariance contribution | 4.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14811803 109 + 27905053 CAGUUGUUGUU---GCUGUUGUUCUAGUUGCAACAGCAACAGGUUAUCCGCAGUGGCUGUCAAACUUCGUGAUAUGCCCAAAUCCGAAUAUAUUUUCUCGUCUAUCCGCAUU .......((((---((((((((.......))))))))))))((....))((.(.((((((((.......))))).))))......(((......)))..........))... ( -30.10) >DroSec_CAF1 4417 105 + 1 CAGUUGUUGCUGUAGCUGUUGUUCUAGUUGCAACAGCAACAGGUUAUCUGCAGUGGCUGUCAAACUUCGUGAUAUGCCCAAAUCCGAAUAUAUUUCCUCGGCUAU------- ((((..(((((((.((((((((.......)))))))).)))((....))))))..))))......((((.(((........))))))).................------- ( -28.60) >DroSim_CAF1 4425 112 + 1 CAGUUGUUGUUGUAGCUGUUGUUCUAGUUGCAACAGCAACAGGUUAUCUGCAGUGGCUGUCAAACUUCGUGAUAUGCCCAAAACCGAAUUUAUUUCCUCGGCUAUUCGCACU ..((((((((((((((((......))))))))))))))))........(((.(.((((((((.......))))).))))....((((..........))))......))).. ( -35.80) >DroEre_CAF1 6181 102 + 1 CAGUUGUUG---------UUGUUCUAGUUGCAACAGCAACAGGUUAUCUGCAGUGGCUGUCAAACUUCGUGAUAUGCCCAAAUCCGAUUUUAUUUCCUCGGCUCUCCGCG-U ..(((((((---------((((.......))))))))))).........((.(.((((((((.......))))).))))....((((..........))))......)).-. ( -27.50) >consensus CAGUUGUUGUU___GCUGUUGUUCUAGUUGCAACAGCAACAGGUUAUCUGCAGUGGCUGUCAAACUUCGUGAUAUGCCCAAAUCCGAAUAUAUUUCCUCGGCUAUCCGCA_U ..((((((((((((((((......)))))))))))))))).........((.(.((((((((.......))))).))))....((((..........))))......))... (-21.80 = -25.80 + 4.00)


| Location | 14,811,872 – 14,811,980 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 87.72 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14811872 108 - 27905053 AAAUUCGGAUUGCAAGCAGAAAACAA------AAAUGUAAAGGCGAUAAAGUGCCGAGCGAAGCAAAGUGCUAAAAUGCGGAUAGACGAGAAAAUAUAUUCGGAUUUGGGCAUA .........((((..((.....(((.------...)))...((((......))))..))...)))).(((((.((((.((((((............)))))).)))).))))). ( -24.60) >DroSec_CAF1 4489 107 - 1 AAAUUCGGAUUGCAAGCAGAAAAUGAAAAUGAAAAUGAAAAGGCGAUAAAGUGCCGAGCGAAGCAAAGUGCUAA-------AUAGCCGAGGAAAUAUAUUCGGAUUUGGGCAUA .........((((..((.....((....))...........((((......))))..))...)))).(((((..-------....(((((........))))).....))))). ( -22.20) >DroSim_CAF1 4497 114 - 1 AAAUUCGGAUUGCAAGCAGAAAAUGAAAAUGAAAAUGAAAAGGCGAUAAAGUGCCGAGCGAAGCAAAGUGCUAAAGUGCGAAUAGCCGAGGAAAUAAAUUCGGUUUUGGGCAUA ........(((((...((....((....)).....)).....)))))...(((((..(((.(((.....)))....)))....(((((((........)))))))...))))). ( -24.60) >consensus AAAUUCGGAUUGCAAGCAGAAAAUGAAAAUGAAAAUGAAAAGGCGAUAAAGUGCCGAGCGAAGCAAAGUGCUAAA_UGCG_AUAGCCGAGGAAAUAUAUUCGGAUUUGGGCAUA ........(((((...((.................)).....)))))...(((((.((...(((.....))).............(((((........)))))..)).))))). (-15.30 = -15.63 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:29 2006