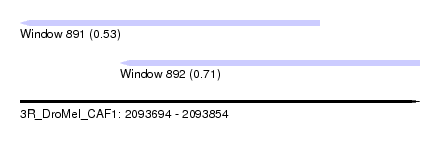
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,093,694 – 2,093,854 |
| Length | 160 |
| Max. P | 0.705756 |

| Location | 2,093,694 – 2,093,814 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.00 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -28.80 |
| Energy contribution | -29.92 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2093694 120 - 27905053 UGCCAUUGUAGCGGGAAAGGCGCUUAUUGUGGGCAAGAUUGCGCUGAUCAUCAGUGCUAUAAUCGGUCUGAAAAAACUUGUGGGUCACGACGGCGGCGAGAAGACCACCUACGAGAUCGU .((((((((((((.((..(((((...(((....)))....)))))..)).....))))))))).))).........(((((((((......(((........).)))))))))))..... ( -34.70) >DroGri_CAF1 245125 120 - 1 UGCCAUUGUCGCUGGCAAGGCGCUGAUUGUGGGCAAAAUAGCGUUGAUCAUCAGCGCCAUCAUUGGAUUGAAGAAGCUCGUCAGUCACGAUGGCGGCGAGAAGACAACCUAUGAGAUUGU .((((((((.((((((..(((((((((.((.(((........))).)).))))))))).(((......)))........)))))).)))))))).........((((.(.....).)))) ( -44.90) >DroWil_CAF1 242238 120 - 1 UGCCAUUGUGGCGGGCAAGGCUCUGAUUGUGGGCAAAAUUGCUCUUAUCAUUAGCGCCAUCAUUGGCCUCAAGAAGCUUGUUAGCCACGAUGGCGGCGAAAAGACCACCUACGAAAUUGU .(((((((((((..((((((((.((((...(((((....)))))..))))..)))((((....)))).........)))))..)))))))))))(((.....).)).............. ( -48.00) >DroMoj_CAF1 249094 120 - 1 AGCCAUUGUGGCCGGCAAGGCGCUGAUCGUGGGCAAGAUCGCGCUGAUCAUCAGCGCCAUCAUCGGGCUGAAGAAGCUGGUCAGCCACGACGGCGGCGAGAAGACCACCUAUGAGAUUGU .(((.(((((((.(((..(((((((((.((.(((........))).)).))))))))).......((((.....)))).))).))))))).)))(((.....).)).............. ( -51.00) >DroAna_CAF1 212973 120 - 1 UGCCAUUGUGGCCGGCAAGGCGUUGAUAGUGGGAAAAAUAGCGCUAAUCAUUAGCGCCAUUAUCGGUUUAAAGAAACUUGUGGGCCACGACGGCGGUGAGAAGACCACAUACGAAAUCGU .(((.((((((((.(((((.(.(((((((((.........((((((.....)))))))))))))))......)...))))).)))))))).)))(((......)))....(((....))) ( -45.00) >DroPer_CAF1 214921 120 - 1 CGCCAUUGUGGCUGGCAAGGCUUUGAUUGUGGGCAAAAUAGCUCUCAUCAUCAGCGCCAUCAUCGGUCUGAAGAAGCUUGUUGGCCACGACGGGGGCGAGAAGACCACCUACGAAAUCGU ((((.(((((((..(((((.(((((((.(.((((......)))).)))).((((.(((......))))))).)))))))))..)))))))....))))............(((....))) ( -46.40) >consensus UGCCAUUGUGGCCGGCAAGGCGCUGAUUGUGGGCAAAAUAGCGCUGAUCAUCAGCGCCAUCAUCGGUCUGAAGAAGCUUGUCAGCCACGACGGCGGCGAGAAGACCACCUACGAAAUCGU .(((.(((((((.((((((((((((((.((((((......)).)).)).))))))))).(((......)))......))))).))))))).)))(((.....).)).............. (-28.80 = -29.92 + 1.12)
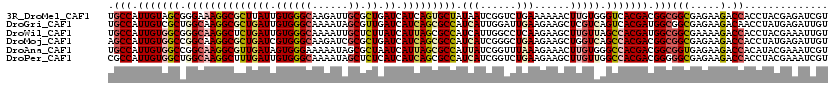
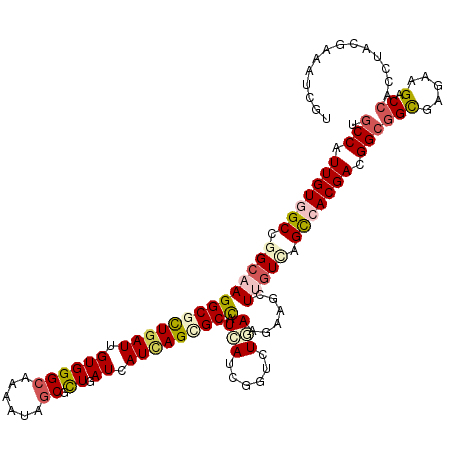
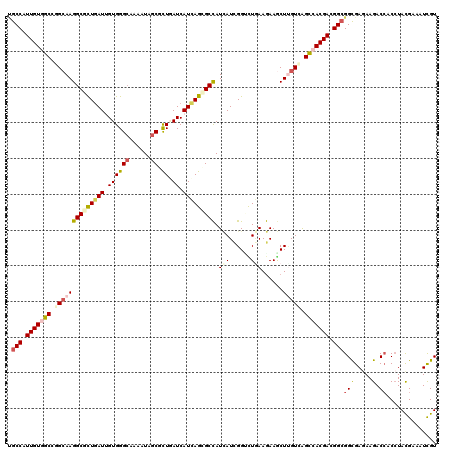
| Location | 2,093,734 – 2,093,854 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -40.85 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2093734 120 - 27905053 UGUUAAAGACCGCUAUUCUAAAGAUGGCAUACCACUCCAUUGCCAUUGUAGCGGGAAAGGCGCUUAUUGUGGGCAAGAUUGCGCUGAUCAUCAGUGCUAUAAUCGGUCUGAAAAAACUUG ......((((((((.((((...(((((((...........)))))))......)))).)))((((.....))))..((((((((((.....))))))...)))))))))........... ( -36.90) >DroVir_CAF1 300996 120 - 1 UGCUAAAGACGGCCAUACUGAAAAUGGCCUACCAUUCGAUUGCCAUUGUGGCUGGCAAGGCGCUGAUUGUGGGCAAAAUCGCGCUGAUCAUCAGCGCCAUCAUUGGGCUAAAGAAGCUGG .(((......((((((.......))))))..(((...((((((((.......))))))(((((((((.((.(((........))).)).))))))))).))..)))........)))... ( -45.40) >DroGri_CAF1 245165 120 - 1 UCCUGAAGACUGCCAUACUUAAGAUGGCCUAUCAUUCGAUUGCCAUUGUCGCUGGCAAGGCGCUGAUUGUGGGCAAAAUAGCGUUGAUCAUCAGCGCCAUCAUUGGAUUGAAGAAGCUCG ((((((....(((((.......((((((..(((....))).)))))).....))))).(((((((((.((.(((........))).)).))))))))).)))..)))............. ( -39.90) >DroWil_CAF1 242278 120 - 1 UGCUAAAAACAGCCAUUUUAAAGAUGGCCUAUCAUUCGAUUGCCAUUGUGGCGGGCAAGGCUCUGAUUGUGGGCAAAAUUGCUCUUAUCAUUAGCGCCAUCAUUGGCCUCAAGAAGCUUG .(((((.....(((((......((((((..(((....))).)))))))))))((((((.((((.......))))....))))))......)))))((((....))))............. ( -38.80) >DroMoj_CAF1 249134 120 - 1 UGCUCAAGACGGCCAUACUGAAGAUGGCCUAUCACUCGAUAGCCAUUGUGGCCGGCAAGGCGCUGAUCGUGGGCAAGAUCGCGCUGAUCAUCAGCGCCAUCAUCGGGCUGAAGAAGCUGG .((((....(((((((......((((((.((((....)))))))))))))))))....(((((((((.((.(((........))).)).)))))))))......))))............ ( -51.70) >DroAna_CAF1 213013 120 - 1 UUCUAAAAACAGCUAUCCUGAAAAUGGCCUACCAUUCGAUUGCCAUUGUGGCCGGCAAGGCGUUGAUAGUGGGAAAAAUAGCGCUAAUCAUUAGCGCCAUUAUCGGUUUAAAGAAACUUG .............((..((((.((((.(((((...(((((.(((.((((.....))))))))))))..))))).......((((((.....)))))))))).))))..)).......... ( -32.40) >consensus UGCUAAAGACAGCCAUACUGAAGAUGGCCUACCAUUCGAUUGCCAUUGUGGCCGGCAAGGCGCUGAUUGUGGGCAAAAUAGCGCUGAUCAUCAGCGCCAUCAUCGGGCUGAAGAAGCUUG .........(.(((((......((((((.............))))))))))).)....(((((((((.((.(((........))).)).)))))))))...................... (-27.48 = -27.12 + -0.36)

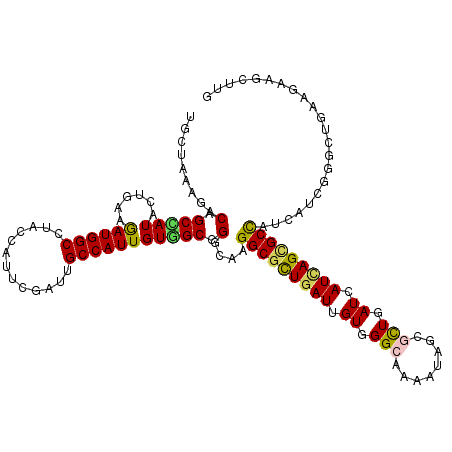
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:06 2006