| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,787,558 – 14,787,675 |
| Length | 117 |
| Max. P | 0.524300 |

| Location | 14,787,558 – 14,787,675 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.95 |
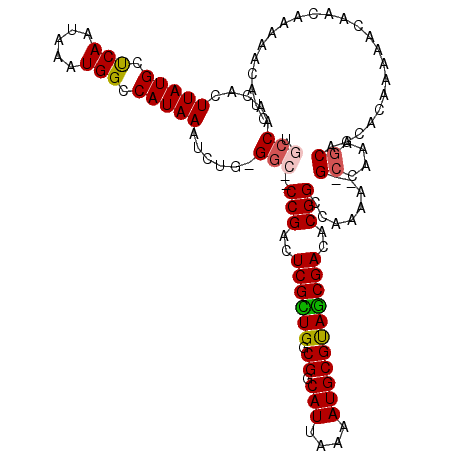
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.62 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
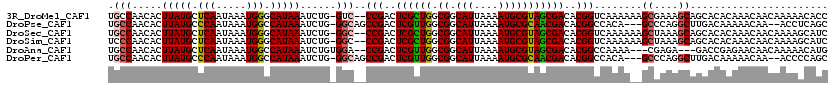
>3R_DroMel_CAF1 14787558 117 - 27905053 UGCCAACACUUAUGCUCAAUAAAUGGGCAUAAAUCUG-GUC--CCGACUCGCUGGCGGCAUUAAAAUGCGUAGCGACACGGUCAAAAAAGCGAAAGCAGCACACAAACAACAAAAACACC .((((....(((((((((.....)))))))))...))-)).--(((..((((((.((.(((....)))))))))))..)))........((....))....................... ( -31.80) >DroPse_CAF1 44184 114 - 1 UGCCAACACUUAUGCCCAAUAAAUGGCCAUAAAUCUG-GGCAGCCGACUCGUUGGCGGCAUUAAAAUGCGCAACGACACGGCCACA---GCCCAGGCUUGACAAAAACAA--ACCUCAGC .(((.....(((((.(((.....))).)))))...((-(((.((((..((((((.((.(((....)))))))))))..))))....---)))))))).............--........ ( -36.10) >DroSec_CAF1 45830 117 - 1 UGCCAACACUUAUGCUCAAUAAAUGGGCAUAAAUCUG-GGC--CCGACUCGCUGGCGGCAUUAAAAUGCGUAGCGACACGGUCAAAAAAGCUAAAGCAGCACACAAACAACAAAAGCAUC .(((.....(((((((((.....))))))))).....-)))--(((..((((((.((.(((....)))))))))))..)))........(((.....))).................... ( -30.80) >DroSim_CAF1 47819 117 - 1 UCCCAACACUUAUGCUCAAUAAAUGGGCAUAAAUCUG-GGC--CCGACUCGCUGGCGGCAUUAAAAUGCGUAGCGACACGGUCAAAAAAGCUAAAGCAGCACACAAACAACAAAAGCAUC .((((....(((((((((.....)))))))))...))-)).--(((..((((((.((.(((....)))))))))))..)))........(((.....))).................... ( -30.50) >DroAna_CAF1 48548 112 - 1 UGCCAACACUUAUGCUCAAUAAAUGGCCAUAAAUCUGUGGA--CCGACUCGUUGGCGGCAUUAAAAUGCGUAGCGACACGGCCAAAA---CGAGA---GACCGAGAACAACAAAAACAUG ..............(((......((((((((....))))).--))).(((((((((.((((....))))((....))...))))..)---)))).---....)))............... ( -25.80) >DroPer_CAF1 44289 114 - 1 UGCCAACACUUAUGCCCAAUAAAUGGCCAUAAAUCUG-GGCAGCCGACUCGUUGGCGGCAUUAAAAUGCGCAACGACACGGCCACA---GCCCAGGCUUGACAAAAACAA--ACCCCAGC .(((.....(((((.(((.....))).)))))...((-(((.((((..((((((.((.(((....)))))))))))..))))....---)))))))).............--........ ( -36.10) >consensus UGCCAACACUUAUGCUCAAUAAAUGGCCAUAAAUCUG_GGC__CCGACUCGCUGGCGGCAUUAAAAUGCGUAGCGACACGGCCAAAA__GCCAAAGCAGCACAAAAACAACAAAAACAUC .(((.....(((((.(((.....))).)))))......)))..(((..((((((.((.(((....)))))))))))..)))........((....))....................... (-18.82 = -18.62 + -0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:08 2006