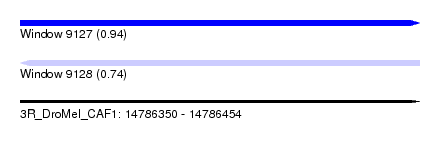
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,786,350 – 14,786,454 |
| Length | 104 |
| Max. P | 0.936146 |

| Location | 14,786,350 – 14,786,454 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.54 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
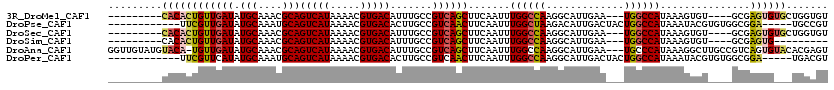
>3R_DroMel_CAF1 14786350 104 + 27905053 ACACCAGCACACUCGC----ACACUUUAUGGCCA---UUCAAUGCCUUGGCCAAAUUGAAGCUGACGGCAAAUGUCACGUUUUAUGACUGCGUUUGCAUAUCAACAGUGUG--------- .......(((((((((----(..(((((((((((---..........))))))...))))).)).))(((((((.((.(((....))))))))))))........))))))--------- ( -29.10) >DroPse_CAF1 42687 103 + 1 ACGGCA-----UCCGCCACACGUAUUUAUGGCCAGUAGUCAAUGUCUUAGCCAAAUUGAAGUUGACGGCAAGUGUCACGUUUUAUGACUGCAUUUGCAUAUCAACGAA------------ ..(((.-----...)))...........((((....((.......))..)))).......(((((..(((((((.((.(((....))))))))))))...)))))...------------ ( -25.50) >DroSec_CAF1 44599 104 + 1 ACACCAGCACACUCGC----ACACUUUAUGGCCA---UUCAAUGCCUUGGCCAAAUUGAAGCUGACGGCAAAUGUCACGUUUUAUGACUGCGUUUGCAUAUCAACAGUGUG--------- .......(((((((((----(..(((((((((((---..........))))))...))))).)).))(((((((.((.(((....))))))))))))........))))))--------- ( -29.10) >DroSim_CAF1 46599 95 + 1 ---------CACUCGC----ACACUUUAUGGCCA---UUCAAUGCCUUGGCCAAAUUGAAGCUGACGGCAAAUGUCACGUUUUAUGACUGCGUUUGCAUAUCAACAGUGUG--------- ---------....(((----((.(((((((((((---..........))))))...))))).(((..(((((((.((.(((....))))))))))))...)))...)))))--------- ( -28.20) >DroAna_CAF1 47164 116 + 1 ACUCGUGUACACUGACGGCAAGCCUUUAUGGGCA---UUCAAUGCCUUGGCCAAAUUGAAGCUGACGGCAAAUGUCACGUUUUAUGACUGCGUUUGCAUAUCAACA-UGUACAUACAACC ....(((((((.((.((((..((((....)))).---((((((((....))...))))))))))...(((((((.((.(((....)))))))))))).......))-)))))))...... ( -35.40) >DroPer_CAF1 42764 103 + 1 ACGUCA-----UCCGCCACACGUAUUUAUGGCCAGUAGUCAAUGCCUUGGCCAAAUUGAAGUUGACGGCAAGUGUCACGUUUUAUGACUGCAUUUGCAUAUGAACGAA------------ .(((((-----(........(((.(((((((((((((.....)))..))))))...))))....)))(((((((.((.(((....))))))))))))..))).)))..------------ ( -26.90) >consensus ACACCA___CACUCGC____ACACUUUAUGGCCA___UUCAAUGCCUUGGCCAAAUUGAAGCUGACGGCAAAUGUCACGUUUUAUGACUGCGUUUGCAUAUCAACAGUGUG_________ .......((((((...............((((((.............)))))).......(.(((..(((((((.((.(((....))))))))))))...))).)))))))......... (-19.28 = -19.54 + 0.25)

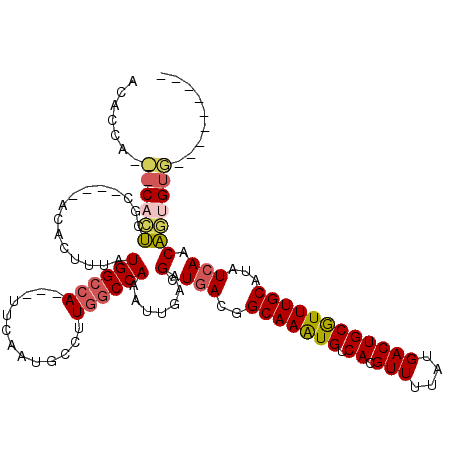
| Location | 14,786,350 – 14,786,454 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14786350 104 - 27905053 ---------CACACUGUUGAUAUGCAAACGCAGUCAUAAAACGUGACAUUUGCCGUCAGCUUCAAUUUGGCCAAGGCAUUGAA---UGGCCAUAAAGUGU----GCGAGUGUGCUGGUGU ---------.(((((((((((..(((((....(((((.....))))).))))).)))))).......((((((..........---))))))...)))))----((.((....)).)).. ( -29.20) >DroPse_CAF1 42687 103 - 1 ------------UUCGUUGAUAUGCAAAUGCAGUCAUAAAACGUGACACUUGCCGUCAACUUCAAUUUGGCUAAGACAUUGACUACUGGCCAUAAAUACGUGUGGCGGA-----UGCCGU ------------...((((((.(((....)))(((((.....))))).......))))))........(((.....((........))((((((......))))))...-----.))).. ( -24.80) >DroSec_CAF1 44599 104 - 1 ---------CACACUGUUGAUAUGCAAACGCAGUCAUAAAACGUGACAUUUGCCGUCAGCUUCAAUUUGGCCAAGGCAUUGAA---UGGCCAUAAAGUGU----GCGAGUGUGCUGGUGU ---------.(((((((((((..(((((....(((((.....))))).))))).)))))).......((((((..........---))))))...)))))----((.((....)).)).. ( -29.20) >DroSim_CAF1 46599 95 - 1 ---------CACACUGUUGAUAUGCAAACGCAGUCAUAAAACGUGACAUUUGCCGUCAGCUUCAAUUUGGCCAAGGCAUUGAA---UGGCCAUAAAGUGU----GCGAGUG--------- ---------((((((((((((..(((((....(((((.....))))).))))).)))))).......((((((..........---))))))...)))))----)......--------- ( -28.60) >DroAna_CAF1 47164 116 - 1 GGUUGUAUGUACA-UGUUGAUAUGCAAACGCAGUCAUAAAACGUGACAUUUGCCGUCAGCUUCAAUUUGGCCAAGGCAUUGAA---UGCCCAUAAAGGCUUGCCGUCAGUGUACACGAGU .......((((((-(((((((..(((((....(((((.....))))).))))).)))))).......((((...((((.....---.(((......))).)))))))))))))))..... ( -31.30) >DroPer_CAF1 42764 103 - 1 ------------UUCGUUCAUAUGCAAAUGCAGUCAUAAAACGUGACACUUGCCGUCAACUUCAAUUUGGCCAAGGCAUUGACUACUGGCCAUAAAUACGUGUGGCGGA-----UGACGU ------------.((((((.......(((((.(((((.....))))).(((((((............))).)))))))))........((((((......)))))))))-----)))... ( -28.80) >consensus _________CACACUGUUGAUAUGCAAACGCAGUCAUAAAACGUGACAUUUGCCGUCAGCUUCAAUUUGGCCAAGGCAUUGAA___UGGCCAUAAAGUCU____GCGAGUG___UGGCGU .........((((((((((((.(((....)))(((((.....))))).......)))))).......((((((.............))))))...............))))))....... (-21.10 = -21.27 + 0.17)
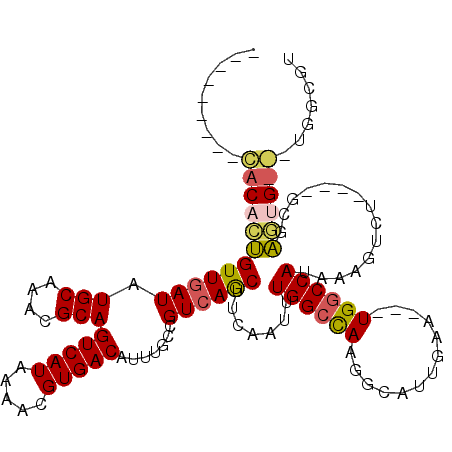
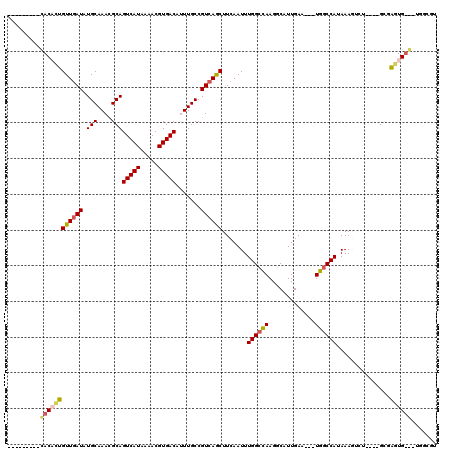
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:06 2006