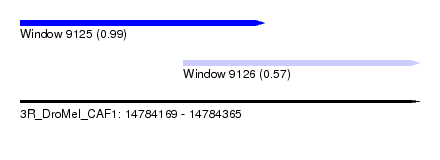
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,784,169 – 14,784,365 |
| Length | 196 |
| Max. P | 0.993171 |

| Location | 14,784,169 – 14,784,289 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -46.12 |
| Consensus MFE | -45.13 |
| Energy contribution | -45.27 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
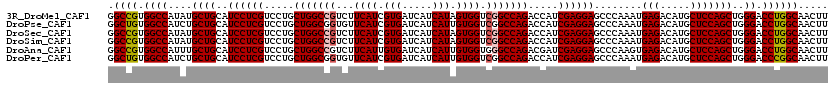
>3R_DroMel_CAF1 14784169 120 + 27905053 GGCCGUGGCCAUAUGCUGCAUCCUCGUCCUGCUGGCCGUCUUCAUCGUGAUCAUCAUAGUGGUCGGCCAGACCAUCGAGGAGCCCAAAUGAGACAUGCUCCAGCUGGGACCUGGCAACUU .((((.((((....((((..((((((.....(((((((...((((..((.....))..)))).))))))).....))))))........(((.....)))))))..)).))))))..... ( -48.60) >DroPse_CAF1 40319 120 + 1 GGCUGUGGCCAUCUGCUGCAUCCUCGUCCUGCUGGCGGUGUUCAUCGUGAUCAUCAUUGUGGUCGGCCAGACCAUCGAGGAGCCCAAAUGAGACAUGCUCCAGCUGGGACCUGGCAACUU (((....)))...((((...((((((.....(((((.(...((((.(((.....))).)))).).))))).....)))))).((((..((.((.....))))..))))....)))).... ( -41.70) >DroSec_CAF1 42485 120 + 1 GGCCGUGGCCAUAUGCUGCAUCCUCGUCCUGCUGGCCGUCUUCAUCGUGAUCAUCAUAGUGGUCGGCCAGACCAUCGAGGAGCCCAAAUGAGACAUGCUCCAGCUGGGACCUGGCAACUU .((((.((((....((((..((((((.....(((((((...((((..((.....))..)))).))))))).....))))))........(((.....)))))))..)).))))))..... ( -48.60) >DroSim_CAF1 44483 120 + 1 GGCCGUGGCCAUAUGCUGCAUCCUCGUCCUGCUGGCCGUCUUCAUCGUGAUCAUCAUAGUGGUCGGCCAGACCAUCGAGGAGCCCAAAUGAGACAUGCUCCAGCUGGGACCUGGCAACUU .((((.((((....((((..((((((.....(((((((...((((..((.....))..)))).))))))).....))))))........(((.....)))))))..)).))))))..... ( -48.60) >DroAna_CAF1 44965 120 + 1 GGCCGUGGCCAUUUGCUGCAUCCUCGUCCUGCUGGCCGUCUUCAUUGUGAUCAUCAUUGUGGUGGGCCAGACGAUCGAGGAGCCCAAGUGAGACAUGCUCCAGCUGGGACCUGGCAACUU .((((.((((....(((((.((((((((...((((((....((((.(((.....))).))))..))))))..)).))))))))....(.(((.....)))))))..)).))))))..... ( -45.30) >DroPer_CAF1 40449 120 + 1 GGCUGUGGCCAUCUGCUGCAUCCUCGUCCUGCUGGCGGUGUUCAUCGUGAUCAUCAUUGUGGUCGGCCAGACCAUCGAGGAGCCCAAAUGAGACAUGCUCCAGCUGGGACCCGGCAACUU .((((.((((....((((..((((((.....(((((.(...((((.(((.....))).)))).).))))).....))))))........(((.....)))))))..)).))))))..... ( -43.90) >consensus GGCCGUGGCCAUAUGCUGCAUCCUCGUCCUGCUGGCCGUCUUCAUCGUGAUCAUCAUAGUGGUCGGCCAGACCAUCGAGGAGCCCAAAUGAGACAUGCUCCAGCUGGGACCUGGCAACUU .((((.((((....((((..((((((.....(((((((...((((.(((.....))).)))).))))))).....))))))........(((.....)))))))..)).))))))..... (-45.13 = -45.27 + 0.14)

| Location | 14,784,249 – 14,784,365 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -19.27 |
| Energy contribution | -19.13 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
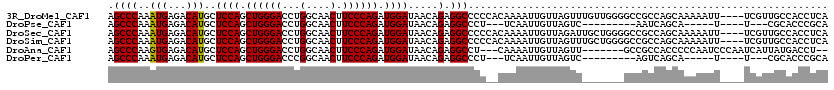
>3R_DroMel_CAF1 14784249 116 + 27905053 AGCCCAAAUGAGACAUGCUCCAGCUGGGACCUGGCAACUUCCCAGAUGGAUAACAGAGGCCCCCACAAAAUUGUUAGUUUGUUGGGGCCGCCAGCAAAAAUU----UCGUUGCCACCUCA .((...(((((((..(((((((.((((((...(....).)))))).)))).......((((((.(((((........))))).))))))....)))....))----)))))))....... ( -41.50) >DroPse_CAF1 40399 96 + 1 AGCCCAAAUGAGACAUGCUCCAGCUGGGACCUGGCAACUUCCCAGAUGGAUAACAGAGGCCCU---UCAAUUGUUAGUC---------AAUCAGCA-----U----U---CGCACCCGCA .((........(((....((((.((((((...(....).)))))).))))((((((((....)---))...))))))))---------.....((.-----.----.---.))....)). ( -25.40) >DroSec_CAF1 42565 116 + 1 AGCCCAAAUGAGACAUGCUCCAGCUGGGACCUGGCAACUUCCCAGAUGGAUAACAGAGGCCCCCACAAAAUUGUUAGAUUGCUGGGGCCGCCAGCAAAAAUU----UCGUUGCCACCUCA .((...(((((((..(((((((.((((((...(....).)))))).)))).......((((((..(((..........)))..))))))....)))....))----)))))))....... ( -37.40) >DroSim_CAF1 44563 116 + 1 AGCCCAAAUGAGACAUGCUCCAGCUGGGACCUGGCAACUUCCCAGAUGGAUAACAGAGGCCCCCACAAAAUUGUUAGUUUGCUGGGGCCGCCAGCAAAAAUU----UCGUUGCCACCUCA .((...(((((((..(((((((.((((((...(....).)))))).)))).......((((((..((((........))))..))))))....)))....))----)))))))....... ( -39.20) >DroAna_CAF1 45045 108 + 1 AGCCCAAGUGAGACAUGCUCCAGCUGGGACCUGGCAACUUCCCAGAUGGAUAACAGAGGCCU---CAAAAUUGUUAGUU-------GCCGCCACCCCCAAUCCCAAUCAUUAUGACCU-- ....(((((((.......((((.((((((...(....).)))))).)))).......(((..---(((.........))-------)..)))..............))))).))....-- ( -23.90) >DroPer_CAF1 40529 96 + 1 AGCCCAAAUGAGACAUGCUCCAGCUGGGACCCGGCAACUUCCCAGAUGGAUAACAGAGGCCCU---UCAAUUGUUAGUC---------AGUCAGCA-----U----U---CGCACCCGCA .((........(((....((((.((((((...(....).)))))).))))((((((((....)---))...))))))))---------.....((.-----.----.---.))....)). ( -24.70) >consensus AGCCCAAAUGAGACAUGCUCCAGCUGGGACCUGGCAACUUCCCAGAUGGAUAACAGAGGCCCC__CAAAAUUGUUAGUU_______GCCGCCAGCAAAAAUU____UCGUUGCCACCUCA .((((..(((...)))..((((.((((((...(....).)))))).)))).....).)))............................................................ (-19.27 = -19.13 + -0.14)
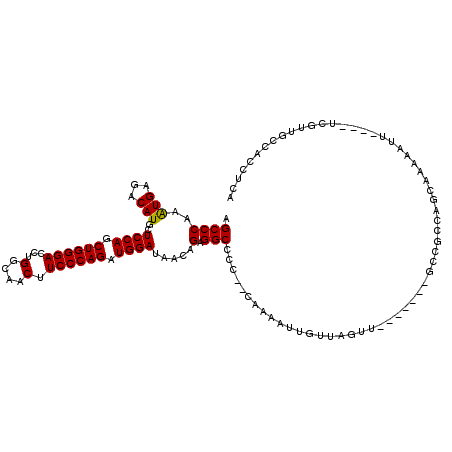
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:04 2006