| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,086,088 – 2,086,200 |
| Length | 112 |
| Max. P | 0.517100 |

| Location | 2,086,088 – 2,086,200 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.59 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -18.55 |
| Energy contribution | -17.67 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
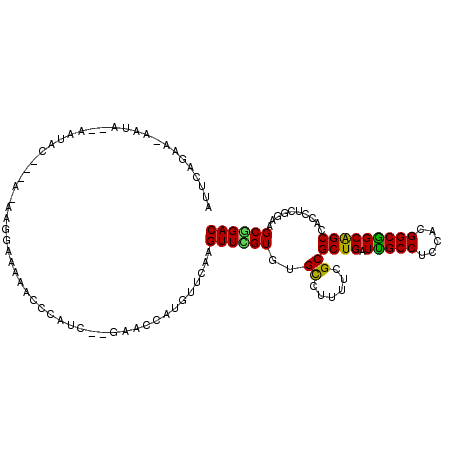
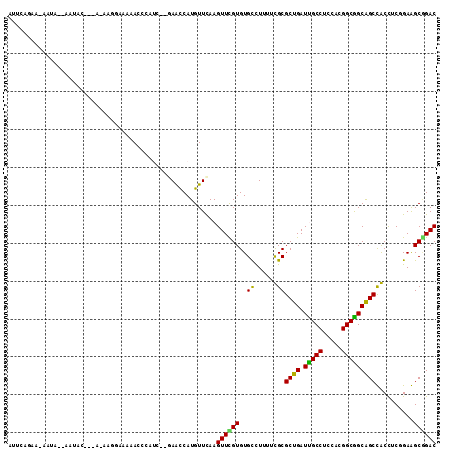
>3R_DroMel_CAF1 2086088 112 + 27905053 AUUCAAAAAAAAAAUAAUACAAAA-AAGGAAAUACCCAUCACGAGCCAUGUUCAAGUUCGUGUGCCUUUUUGCGCUGAUUGCCUCCACGGCGGCGGCUACCUCGGAAGCGGAC .....................(((-((((..........(((((((.........)))))))..)))))))..((((.(((((.....)))))))))..((.(....).)).. ( -28.40) >DroPse_CAF1 205446 95 + 1 GAGUUGAUCAAUU--AAU-------------CAACCAAUC---AAUCAUGUUCAAGUUUGUGUGCUUAUUCGCGCUGAUCGCCUCAACGGCGGCAGCCGCCUCCGAGGCAGAC (.((((((.....--.))-------------)))))....---................((.(((((....((((((.(((((.....)))))))).))).....))))).)) ( -27.70) >DroEre_CAF1 230352 109 + 1 AUUCAGAG-AAUA--AAUACGAGC-AAGGAAAUACCCAUCCGGAGCCAUGUUCAAGUUCGUGUGCCUUUUUGCGCUGAUUGCCUCCACGGCGGCGGCUACCUCGGAAGCGGAC ...(((((-(...--.((((((((-..(((........))).((((...))))..))))))))...)))))).((((.(((((.....)))))))))..((.(....).)).. ( -33.00) >DroYak_CAF1 232775 110 + 1 AUUCAAAA-AAUA--AAUACGAAAAAAGGAAAUACCCAACCCGAGCCAUGUUCAAGUUUGUGUGCCUUUUUGCGCUGAUUGCCUCCACGGCGGCAGCUACCUCGGAAGCGGAC ...(((((-(...--.(((((((....((......)).....((((...))))...)))))))...)))))).((((.(((((.....)))))))))..((.(....).)).. ( -27.10) >DroAna_CAF1 205771 97 + 1 ---CAGAG-AAAA----CUC---A-AAUCAAAAACCUA----AAAUCAUGUUUAAGUUCGUGUGUUUGUUCGCGCUGAUUGCCUCCACGGCAGCAGCCGCUUCUGAGGCCGAC ---(((((-....----..(---(-(((((..(((.((----((......)))).)))..)).)))))...((((((.(((((.....)))))))).))))))))........ ( -23.80) >DroPer_CAF1 207179 95 + 1 GAGUUGAUCAAUU--AAU-------------CAACCAAUC---AAUCAUGUUCAAGUUUGUGUGCUUAUUCGCGCUGAUCGCCUCAACGGCGGCAGCCGCCUCCGAGGCAGAC (.((((((.....--.))-------------)))))....---................((.(((((....((((((.(((((.....)))))))).))).....))))).)) ( -27.70) >consensus AUUCAGAA_AAUA__AAUAC___A_AAGGAAAAACCCAUC__GAACCAUGUUCAAGUUCGUGUGCCUUUUCGCGCUGAUUGCCUCCACGGCGGCAGCCACCUCGGAAGCGGAC .......................................................((((((..((......))((((.(((((.....)))))))))..........)))))) (-18.55 = -17.67 + -0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:02 2006