
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,771,599 – 14,771,752 |
| Length | 153 |
| Max. P | 0.581265 |

| Location | 14,771,599 – 14,771,712 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -17.87 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
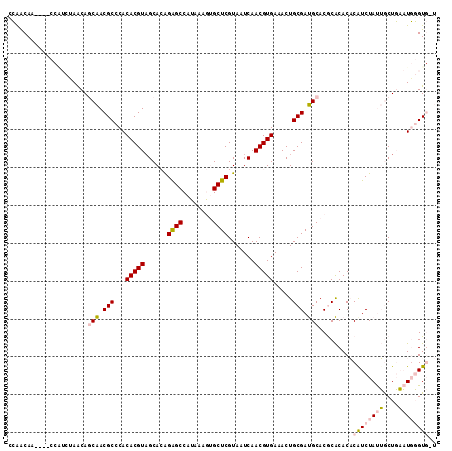
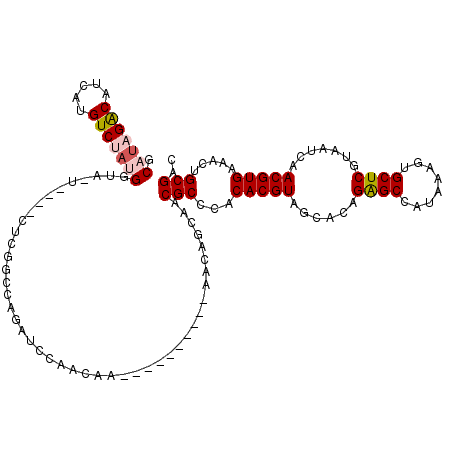
>3R_DroMel_CAF1 14771599 113 - 27905053 CCAACAACCAACCAUCUAACAGCAACGCCCACACGUAGCACAGAGCCAUAAAGUGCUCGUAAUCAACGUGAAACUGCGAUGCACGCACACACAUCUAUUGCUGAAUGGGUGAU .........................((((((....(((((.(((........(((((((((.((.....))...))))).)))).........)))..)))))..)))))).. ( -27.03) >DroPse_CAF1 28909 100 - 1 AGGGCAA----------AGCAACGACGCCCACACGUAGCAGAGGGCCAUAAAAGGCUCGUAAUCAACGUGAAACUGCGACGCACACGCACCCAUCUACGACUCCUUUCGU--- .((((..----------.........)))).(((((......(((((......))))).......)))))....((((.......)))).......((((......))))--- ( -25.52) >DroSim_CAF1 32093 113 - 1 CCAACAACCAACCAUCUAACAGCAACGCCCACACGUAGCACGGAGCCAUAAAGUGCUCGUAAUCAACGUGAAACUGCGAUGCACGCACACACAUCUAUUGCUGAAUGGGUGAU ............((((((.((((((.......(((.(((((...........)))))))).......(((....((((.....))))..))).....))))))..)))))).. ( -27.70) >DroEre_CAF1 30738 109 - 1 CCAACAA----CCAUCCAACAGCAACGCCCACACGUCGCACGGAGCCAUAAAGUGCUCGUAAUCAACGUGAAACUGCGAUGCACGCACACACGUCUAUUGCUGAAUGGGUGUU .......----.((((((...((.(((......))).))...(((((.....).))))(((((..(((((....((((.....))))..)))))..)))))....)))))).. ( -30.00) >DroYak_CAF1 39655 109 - 1 CCAACAA----CCGUCUAACAGCAACGCCCACACGUAGCACGGAGCCAUAAAGUGCUCGUAAUCAACGUGAAACUGCGAUGCACGCACACACGUCUAUUGCUGAAUGGGUGUU .......----............((((((((...(.(((((...........))))))(((((..(((((....((((.....))))..)))))..)))))....)))))))) ( -30.80) >DroPer_CAF1 28965 100 - 1 AGGGCAA----------AGCAACGACGCCCACACGUAGCAGAGGGCCAUAAAAGGCUCGUAAUCAACGUGAAACUGCGACGCACACGCACCCAUCUACGACUCCUUUCGU--- .((((..----------.........)))).(((((......(((((......))))).......)))))....((((.......)))).......((((......))))--- ( -25.52) >consensus CCAACAA____CCAUCUAACAGCAACGCCCACACGUAGCACAGAGCCAUAAAGUGCUCGUAAUCAACGUGAAACUGCGAUGCACGCACACACAUCUAUUGCUGAAUGGGUG_U .....................(((.(((...(((((......((((........)))).......))))).....))).))).........((((((((....)))))))).. (-17.87 = -18.20 + 0.34)


| Location | 14,771,632 – 14,771,752 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -18.73 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
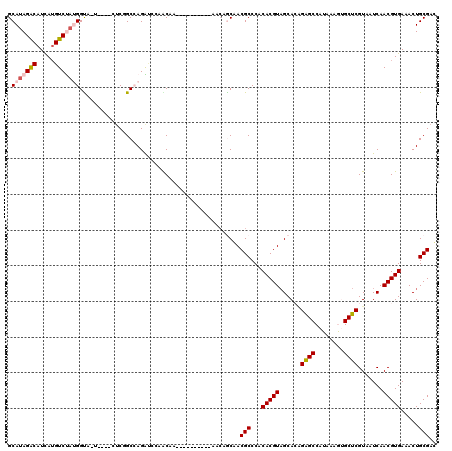
>3R_DroMel_CAF1 14771632 120 - 27905053 GCAUAGACAUCAUGUCUAUGGUCAUACUACUCGGCCAGAUCCAACAACCAACCAUCUAACAGCAACGCCCACACGUAGCACAGAGCCAUAAAGUGCUCGUAAUCAACGUGAAACUGCGAU ((((((((.....))))))((((.........)))).........................))..(((...(((((......(((((.....).)))).......))))).....))).. ( -25.72) >DroPse_CAF1 28939 104 - 1 GCAUAGACAUUAGGUCCAAGGUA------GUCAGCAAGGCAGGGCAA----------AGCAACGACGCCCACACGUAGCAGAGGGCCAUAAAAGGCUCGUAAUCAACGUGAAACUGCGAC .....(((.....)))....(((------((..........((((..----------.........)))).(((((......(((((......))))).......)))))..)))))... ( -27.02) >DroEre_CAF1 30771 116 - 1 GCAUAGACAUCAUGUCUAUGGCCGUACUACUCGGCCAGAUCCAACAA----CCAUCCAACAGCAACGCCCACACGUCGCACGGAGCCAUAAAGUGCUCGUAAUCAACGUGAAACUGCGAU ((((((((.....))))))(((((.......)))))...........----..........))..(((...(((((.(.(((.((((.....).))))))...).))))).....))).. ( -30.50) >DroYak_CAF1 39688 116 - 1 GCAUAGACAUCAUGUCUAUGGUCUUACUACUCGGCCAGAUCCAACAA----CCGUCUAACAGCAACGCCCACACGUAGCACGGAGCCAUAAAGUGCUCGUAAUCAACGUGAAACUGCGAU ((((((((.....))))))((((.........))))...........----..........))..........((((((((((((((.....).))))(....)..))))...))))).. ( -25.40) >DroAna_CAF1 32857 99 - 1 GCAUAGGCAUCAUGUCUUUGGCAGU------C-GCCAGGACAGGC--------------GAGCAACGCCCACACGUAGCACUGAGCCAUAAAGUGCUCGUAAUCAACGUGAAACUGCGAC ((..((((.....))))...)).((------(-((.((....(((--------------(.....))))..(((((((((((.........))))))........)))))...))))))) ( -31.70) >DroPer_CAF1 28995 104 - 1 GCAUAGACAUUAUGUCCAAGGUA------GUCAGCAAGGCAGGGCAA----------AGCAACGACGCCCACACGUAGCAGAGGGCCAUAAAAGGCUCGUAAUCAACGUGAAACUGCGAC .....(((.....)))....(((------((..........((((..----------.........)))).(((((......(((((......))))).......)))))..)))))... ( -27.42) >consensus GCAUAGACAUCAUGUCUAUGGUA_U____CUCGGCCAGAUCCAACAA__________AACAGCAACGCCCACACGUAGCACAGAGCCAUAAAGUGCUCGUAAUCAACGUGAAACUGCGAC .(((((((.....))))))).............................................(((...(((((......((((........)))).......))))).....))).. (-18.73 = -19.20 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:50 2006