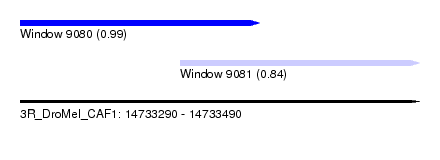
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,733,290 – 14,733,490 |
| Length | 200 |
| Max. P | 0.985807 |

| Location | 14,733,290 – 14,733,410 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -42.47 |
| Consensus MFE | -39.74 |
| Energy contribution | -39.55 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14733290 120 + 27905053 GAAGAAGCUCGGCGAGGACAACAGCGUGUUCGCGCAGGCGCUCGUCGAAUUCGGCGAAGCGCUGAAACAGAUGGCCGACGUCAAGUAUUCGCUGGACGACAACAUCAAGCAGAACUUUUU .((((((.((.(((((.((......)).)))))((.(((((((((((....))))).))))))......((((.....((((.(((....))).))))....))))..)).)).)))))) ( -44.90) >DroPse_CAF1 3029 120 + 1 UAAGAAGCUCGGCGAGGACAACAGCGUGUUUGCGCAGGCGCUCGUCGAGUUUGGCGAGUCGCUGAAACAGAUGGCCGACGUCAAGUACUCGCUGGACGACAAUAUCAAGCAGAACUUUUU ......(((((((((((....(.((((....)))).).).))))))))(((..((((((.(((......((((.....)))).)))))))))..)))...........)).......... ( -39.80) >DroGri_CAF1 304 120 + 1 UAAGAAGCUCGGCGAGGACAACAGCGUGUUUGCGCAGGCGCUCGUCGAAUUCGGCGAAGCGCUGAAACAAAUGGCCGACGUCAAGUAUUCGCUGGACGACAACAUCAAGCAGAACUUUUU ......(((((((..........((((....)))).(((((((((((....))))).))))))..........)))))((((.(((....))).))))..........)).......... ( -40.70) >DroWil_CAF1 304 120 + 1 UAAGAAGCUCGGCGAGGACAACAGCGUGUUCGCGCAGGCGCUCGUCGAAUUCGGCGAGGCGCUGAAGCAGAUGGCCGACGUCAAGUAUUCGCUGGACGACAACAUCAAGCAGAACUUUUU .((((((.((.(((((.((......)).)))))((.(((((((((((....)))))).)))))......((((.....((((.(((....))).))))....))))..)).)).)))))) ( -44.70) >DroAna_CAF1 304 120 + 1 GAAGAAGCUCGGCGAGGACAACAGCGUGUUCGCGCAGGCGCUCGUCGAGUUCGGCGAGGCGCUAAAACAGAUGGCCGACGUCAAGUAUUCGCUGGACGACAACAUCAAGCAGAACUUUUU .((((((.((.(((((.((......)).)))))((.(((((((((((....)))))).)))))......((((.....((((.(((....))).))))....))))..)).)).)))))) ( -44.90) >DroPer_CAF1 3060 120 + 1 UAAGAAGCUCGGCGAGGACAACAGCGUGUUUGCGCAGGCGCUCGUCGAGUUUGGCGAGUCGCUGAAACAGAUGGCCGACGUCAAGUACUCGCUGGACGACAAUAUCAAGCAGAACUUUUU ......(((((((((((....(.((((....)))).).).))))))))(((..((((((.(((......((((.....)))).)))))))))..)))...........)).......... ( -39.80) >consensus UAAGAAGCUCGGCGAGGACAACAGCGUGUUCGCGCAGGCGCUCGUCGAAUUCGGCGAGGCGCUGAAACAGAUGGCCGACGUCAAGUAUUCGCUGGACGACAACAUCAAGCAGAACUUUUU .((((((.((.(((((.((......)).)))))((.(((((((((((....)))))).)))))......((((.....((((.(((....))).))))....))))..)).)).)))))) (-39.74 = -39.55 + -0.19)



| Location | 14,733,370 – 14,733,490 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -33.46 |
| Energy contribution | -33.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14733370 120 + 27905053 UCAAGUAUUCGCUGGACGACAACAUCAAGCAGAACUUUUUGGAGCCACUGCAUCAUAUGCAGACCAAAGACCUCAAGGAGGUAAUGCAUCAUCGCAAGAAGCUGCAGGGCCGGCGGCUAG (((((..((((((.((........)).))).)))...)))))((((.((((((...)))))).((.....((....)).(((..((((.(.((....)).).))))..))))).)))).. ( -35.80) >DroVir_CAF1 384 120 + 1 UCAAGUAUUCGCUGGACGACAACAUCAAGCAGAACUUUUUGGAGCCUCUGCAUCAUAUGCAGACCAAAGACCUAAAGGAGGUCAUGCACCAUCGAAAGAAGCUGCAGGGCCGGCGUCUCG .........((((((.(((.....))..((((..((((((((.((.(((((((...))))))).....(((((.....)))))..)).)))..)))))...)))).)..))))))..... ( -37.40) >DroPse_CAF1 3109 120 + 1 UCAAGUACUCGCUGGACGACAAUAUCAAGCAGAACUUUUUGGAGCCCCUGCAUCAUAUGCAGAUGAAAGACCUCAAGGAGGUUAUGCACCAUCGCAAGAAGCUGCAGGGCCGGCGCCUCG .........((((((.(((.....))..((((.......(((.((..((((((...))))))......((((((...))))))..)).)))((....))..)))).)..))))))..... ( -34.70) >DroWil_CAF1 384 120 + 1 UCAAGUAUUCGCUGGACGACAACAUCAAGCAGAACUUUUUGGAGCCACUGCAUCAUAUGCAGACCAAAGACCUCAAAGAGGUAAUGCACCAUCGAAAGAAACUGCAGGGUCGGCGCCUCG ..((((.(..(((.((........)).)))..)))))...((.(((..(((((...)))))((((....(((((...)))))..((((...((....))...)))).))))))).))... ( -34.10) >DroMoj_CAF1 384 120 + 1 UCAAGUAUUCGCUGGACGACAACAUCAAGCAGAACUUUUUGGAGCCCCUGCAUCAUAUGCAGACCAAAGACCUUAAGGAGGUCAUGCACCAUCGAAAGAAGCUGCAGGGCCGGCGUCUCG .........((((((.(((.....))..((((..((((((((.((..((((((...))))))......((((((...))))))..)).)))..)))))...)))).)..))))))..... ( -36.40) >DroPer_CAF1 3140 120 + 1 UCAAGUACUCGCUGGACGACAAUAUCAAGCAGAACUUUUUGGAGCCCCUGCAUCAUAUGCAGAUGAAAGACCUCAAGGAGGUUAUGCACCAUCGCAAGAAGCUGCAGGGCCGGCGCCUCG .........((((((.(((.....))..((((.......(((.((..((((((...))))))......((((((...))))))..)).)))((....))..)))).)..))))))..... ( -34.70) >consensus UCAAGUAUUCGCUGGACGACAACAUCAAGCAGAACUUUUUGGAGCCCCUGCAUCAUAUGCAGACCAAAGACCUCAAGGAGGUAAUGCACCAUCGAAAGAAGCUGCAGGGCCGGCGCCUCG .........((((((.(((.....))..((((.......(((.((..((((((...))))))......((((((...))))))..)).)))((....))..)))).)..))))))..... (-33.46 = -33.47 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:22 2006