
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,729,229 – 14,729,328 |
| Length | 99 |
| Max. P | 0.801599 |

| Location | 14,729,229 – 14,729,328 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -17.14 |
| Energy contribution | -17.12 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14729229 99 + 27905053 UAUACUCUUAGUUGGGCAGU-----UGCGGCUGGAUUUCAUUCUGUGC-UCGAAAUUACCAUUUU--------UUGUUUUGCUGCUGCUGCCUGAAUUUCAAAUGCCGCAACU ....(((......))).(((-----((((((((((.((((..(.((((-.((((((.........--------..))))))..)).)).)..)))).))))...))))))))) ( -26.20) >DroSim_CAF1 153626 107 + 1 UGUACUCUUAGUUGGGCAGU-----UGCGGCUGGAUUUCAUUCUGUGC-UCGAAAUUACCAUUUUUGGUAAUUUUGUUUUGCUGCCGCUGCCUGAAUUUCAAAUGCCGCAACU .........(((((((((((-----.(((((.(((......)))..((-.(((((((((((....)))))))))))....)).))))).)).((.....))..)))).))))) ( -36.00) >DroEre_CAF1 146549 91 + 1 UGAACUCUUAGUUGGGCAGU-----UGCGGCUGGAUUUCAUUCUGUGC-ACGGAAUUACCAUUUU--------UU-----G---CUGCCGCCUGAAUUUCAAAUGCCGCAACU ....(((......))).(((-----((((((((((.((((..(.((((-(..((((....)))).--------.)-----)---).)).)..)))).))))...))))))))) ( -26.80) >DroYak_CAF1 156033 91 + 1 UGUACUCUUAGUUGGGCAGU-----UGCGGCUGGAUUUCAUUCUGUGC-UCGGAAUUACCAUUUU--------UU-----G---CUGCCGCCUGAAUUUCAAAUGCCGCAACU ....(((......))).(((-----((((((((((.((((...((.((-.(((((........))--------))-----)---..))))..)))).))))...))))))))) ( -24.80) >DroAna_CAF1 149711 96 + 1 UAUAUAUUUAGUUGGGCAGC-CGAAUGCGGCGGGAUUUUAUUUUGUGCCUCGGCAAUACCAUUUU--------UU-----G---CUGCUGCGUGAAUUUCAAAUGCCGCAACU .........(((((((((((-((....))))..........((..(((..((((((.........--------))-----)---)))..)))..)).......)))).))))) ( -29.90) >DroPer_CAF1 139333 97 + 1 UAUAUAUUUAGUUGGGCAGCCCGAAUGCGGCUGGAUUUUAUUUUGUAC-UCUGGAUUUUCAUUUUU-------UU-----A---CUGCUGCGUGAAUUUCAAAUGCCACAACU .........((((((((((((.......)))((((.(((((...(((.-...(((........)))-------..-----.---.)))...))))).))))..)))).))))) ( -18.90) >consensus UAUACUCUUAGUUGGGCAGU_____UGCGGCUGGAUUUCAUUCUGUGC_UCGGAAUUACCAUUUU________UU_____G___CUGCUGCCUGAAUUUCAAAUGCCGCAACU .........(((((((((........(((((.(((......))).......((.....))..........................))))).((.....))..)))).))))) (-17.14 = -17.12 + -0.03)


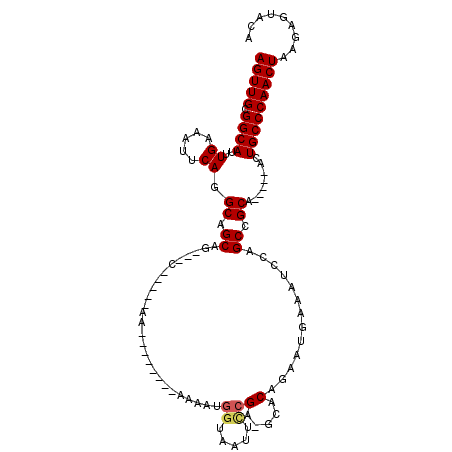
| Location | 14,729,229 – 14,729,328 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -23.31 |
| Consensus MFE | -12.48 |
| Energy contribution | -12.65 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14729229 99 - 27905053 AGUUGCGGCAUUUGAAAUUCAGGCAGCAGCAGCAAAACAA--------AAAAUGGUAAUUUCGA-GCACAGAAUGAAAUCCAGCCGCA-----ACUGCCCAACUAAGAGUAUA (((((.((((.(((.......(((.((....)).......--------....(((..((((((.-.(...)..)))))))))))).))-----).)))))))))......... ( -22.20) >DroSim_CAF1 153626 107 - 1 AGUUGCGGCAUUUGAAAUUCAGGCAGCGGCAGCAAAACAAAAUUACCAAAAAUGGUAAUUUCGA-GCACAGAAUGAAAUCCAGCCGCA-----ACUGCCCAACUAAGAGUACA (((((((((.......((((..((.((....))....(.(((((((((....))))))))).).-))...))))........))))))-----)))................. ( -30.66) >DroEre_CAF1 146549 91 - 1 AGUUGCGGCAUUUGAAAUUCAGGCGGCAG---C-----AA--------AAAAUGGUAAUUCCGU-GCACAGAAUGAAAUCCAGCCGCA-----ACUGCCCAACUAAGAGUUCA (((((.((((.(((.....)))(((((.(---(-----(.--------....(((.....))))-))......((.....))))))).-----..)))))))))......... ( -24.20) >DroYak_CAF1 156033 91 - 1 AGUUGCGGCAUUUGAAAUUCAGGCGGCAG---C-----AA--------AAAAUGGUAAUUCCGA-GCACAGAAUGAAAUCCAGCCGCA-----ACUGCCCAACUAAGAGUACA (((((.((((.(((.....)))(((((..---.-----..--------....(((..((((...-.....)))).....)))))))).-----..)))))))))......... ( -22.60) >DroAna_CAF1 149711 96 - 1 AGUUGCGGCAUUUGAAAUUCACGCAGCAG---C-----AA--------AAAAUGGUAUUGCCGAGGCACAAAAUAAAAUCCCGCCGCAUUCG-GCUGCCCAACUAAAUAUAUA (((((.((((((((...(((..((....)---)-----..--------.....((.....)))))...))))..........((((....))-)))))))))))......... ( -23.10) >DroPer_CAF1 139333 97 - 1 AGUUGUGGCAUUUGAAAUUCACGCAGCAG---U-----AA-------AAAAAUGAAAAUCCAGA-GUACAAAAUAAAAUCCAGCCGCAUUCGGGCUGCCCAACUAAAUAUAUA (((((.(((.((((..((((((......)---)-----..-------.....((......))))-)).)))).........((((.......))))))))))))......... ( -17.10) >consensus AGUUGCGGCAUUUGAAAUUCAGGCAGCAG___C_____AA________AAAAUGGUAAUUCCGA_GCACAGAAUGAAAUCCAGCCGCA_____ACUGCCCAACUAAGAGUACA (((((.((((..((.....)).((.((..........................((.....))(.....).............)).))........)))))))))......... (-12.48 = -12.65 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:18 2006