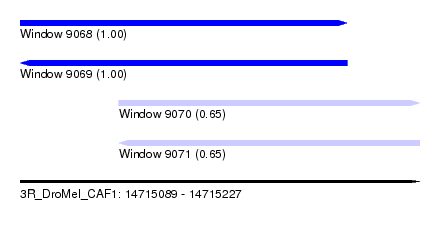
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
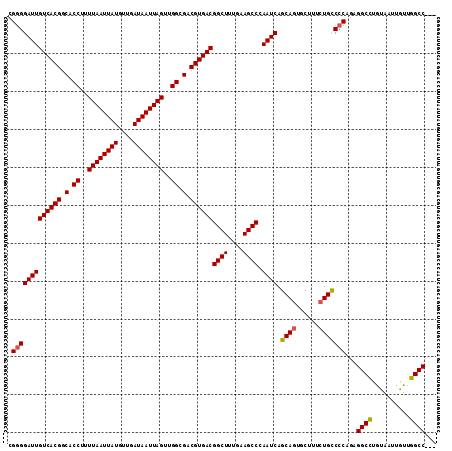
| Location | 14,715,089 – 14,715,227 |
| Length | 138 |
| Max. P | 0.999983 |

| Location | 14,715,089 – 14,715,202 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -35.82 |
| Energy contribution | -36.62 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.92 |
| SVM decision value | 5.00 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
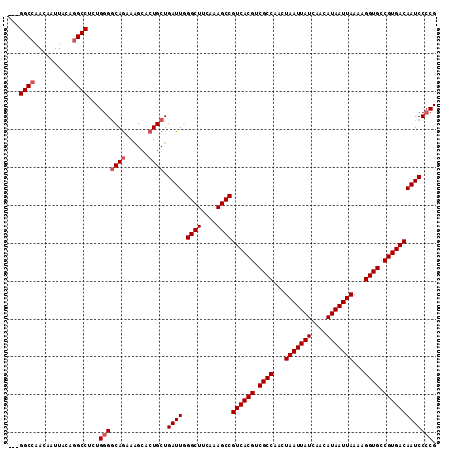
>3R_DroMel_CAF1 14715089 113 + 27905053 GUUGGCCAACAAUUACAAGCCUCUGGGACAGAAAGCACUGCUGAUUGGGCUUCAAAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAAGGUGCCGUGACAAUCCCCG ...(((............)))...(((((((..(((...)))..)))((((....))))((((((.((((...(((((((....)))))))...)))).))))))..)))).. ( -33.60) >DroSec_CAF1 133768 109 + 1 ---GGCCAACAAUUACAGGCCUCUGGGGCAGAAAGCACUGCUGAUUGGGCUUCAAAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAAGGUGCCGUGACAAUCC-CG ---((((..........))))...(((((((......))))......((((....))))((((((.((((...(((((((....)))))))...)))).))))))...))-). ( -39.10) >DroSim_CAF1 138713 110 + 1 ---GGCCAACAAUUACAGGCCUUUGGGGCAGAAAGCACUGCUGAUUGGGCUUCAAAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAAGGUGCCGUGACAAUCCCCG ---((((..........))))...(((((((......)))).((((.((((....))))((((((.((((...(((((((....)))))))...)))).))))))))))))). ( -40.80) >DroEre_CAF1 128873 110 + 1 ---GGCCUAUAAUUAUAGGCCUCUGGGGCAGAAGGCACUGCUGAUUUGGCUUCAAAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAAGGUGCCGUGACAAUCCCCG ---(((((((....)))))))...(((((((......)))).((((.((((....))))((((((.((((...(((((((....)))))))...)))).))))))))))))). ( -44.60) >DroYak_CAF1 135073 110 + 1 ---GGCCAACAAUUACAGGCCUCUGGGGCAAAAAGGAGUGCUGAUUUGGCUUCAAAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAAGGUGCCGUGACAAUCCCCG ---((((..........))))...((((...................((((....))))((((((.((((...(((((((....)))))))...)))).))))))...)))). ( -37.40) >consensus ___GGCCAACAAUUACAGGCCUCUGGGGCAGAAAGCACUGCUGAUUGGGCUUCAAAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAAGGUGCCGUGACAAUCCCCG ...((((..........))))...(((((((......)))).((((.((((....))))((((((.((((...(((((((....)))))))...)))).))))))))))))). (-35.82 = -36.62 + 0.80)

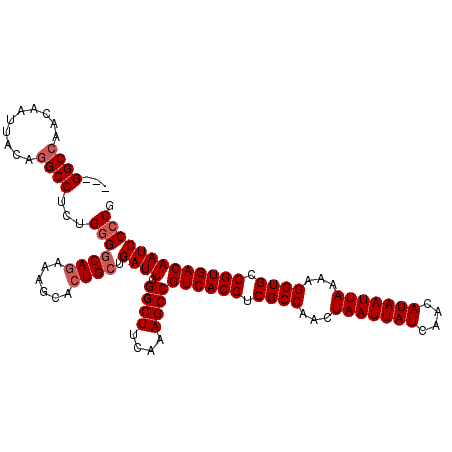
| Location | 14,715,089 – 14,715,202 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -38.70 |
| Energy contribution | -38.78 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.96 |
| SVM decision value | 5.32 |
| SVM RNA-class probability | 0.999983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14715089 113 - 27905053 CGGGGAUUGUCACGGCACCUUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUUUGAAGCCCAAUCAGCAGUGCUUUCUGUCCCAGAGGCUUGUAAUUGUUGGCCAAC .(((((((((((((.(.((..((((((((....))))))))..)).).))))))((((....)))).)))).((((......)))))))...((((..........))))... ( -38.20) >DroSec_CAF1 133768 109 - 1 CG-GGAUUGUCACGGCACCUUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUUUGAAGCCCAAUCAGCAGUGCUUUCUGCCCCAGAGGCCUGUAAUUGUUGGCC--- .(-((...((((((.(.((..((((((((....))))))))..)).).))))))((((....))))......((((......)))))))...((((..........))))--- ( -38.50) >DroSim_CAF1 138713 110 - 1 CGGGGAUUGUCACGGCACCUUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUUUGAAGCCCAAUCAGCAGUGCUUUCUGCCCCAAAGGCCUGUAAUUGUUGGCC--- .(((((((((((((.(.((..((((((((....))))))))..)).).))))))((((....)))).)))).((((......)))))))...((((..........))))--- ( -41.10) >DroEre_CAF1 128873 110 - 1 CGGGGAUUGUCACGGCACCUUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUUUGAAGCCAAAUCAGCAGUGCCUUCUGCCCCAGAGGCCUAUAAUUAUAGGCC--- .(((((((((((((.(.((..((((((((....))))))))..)).).))))))((((....)))).)))).((((......)))))))...(((((((....)))))))--- ( -45.90) >DroYak_CAF1 135073 110 - 1 CGGGGAUUGUCACGGCACCUUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUUUGAAGCCAAAUCAGCACUCCUUUUUGCCCCAGAGGCCUGUAAUUGUUGGCC--- .(((((((((((((.(.((..((((((((....))))))))..)).).))))))((((....)))).)))).(((........))))))...((((..........))))--- ( -38.40) >consensus CGGGGAUUGUCACGGCACCUUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUUUGAAGCCCAAUCAGCAGUGCUUUCUGCCCCAGAGGCCUGUAAUUGUUGGCC___ .(((((((((((((.(.((..((((((((....))))))))..)).).))))))((((....)))).)))).((((......)))))))...((((..........))))... (-38.70 = -38.78 + 0.08)

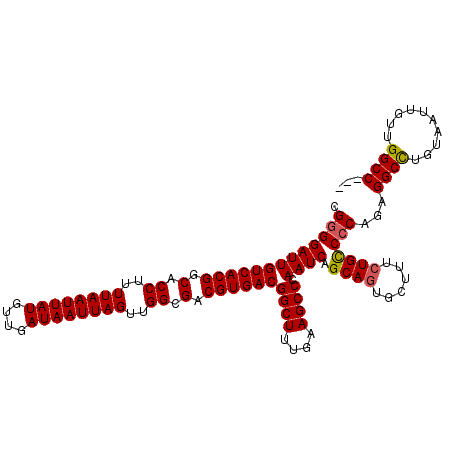
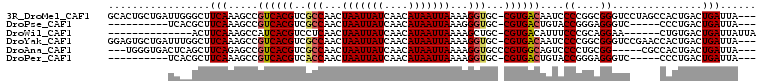
| Location | 14,715,123 – 14,715,227 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -13.86 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14715123 104 + 27905053 GCACUGCUGAUUGGGCUUCAAAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAAGGUGC-CGUGACAAUCCCCGGCGGGUCCUAGCCACUGACUGAUUA--- ...((((((....((((....))))((((((.((((...(((((((....)))))))...)))).-))))))......))))))(((.........)))......--- ( -32.90) >DroPse_CAF1 123589 89 + 1 ----------UCACGCUUCAAAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAGGGUGC-CGUGACUGUACCGGGAGGGUC-----CCCUGACUGAUUA--- ----------....(((....))).((((((.((((...(((((((....)))))))...)))).-)))))).....(((.(((...-----.)))..)))....--- ( -26.40) >DroWil_CAF1 151076 87 + 1 --------------ACUUCAAAGCCAUCACGUCCUCAACUAAUUAUCAACAUAAUUAAAAGCUGC-CGUGACAUUUCCCGCAGGAA------CUGUGACUGAUUAUUA --------------...........((((.(((..((..(((((((....)))))))....((((-.(.((....))).))))...------.)).)))))))..... ( -11.80) >DroYak_CAF1 135104 104 + 1 GGAGUGCUGAUUUGGCUUCAAAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAAGGUGC-CGUGACAAUCCCCGGCGGGUCCGAACCACUGACUGAUUA--- (((.(((((....((((....))))((((((.((((...(((((((....)))))))...)))).-))))))......)))).).))).................--- ( -32.80) >DroAna_CAF1 134192 97 + 1 ---UGGGUGACUCAGCUUCAGAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAAGGUGCCCGUGGCAGUCCCCUGCGG-----CGCCACUGACUGAUUA--- ---..(....)((((..((((.(((..((((.((((...(((((((....)))))))...))))..))))((((....))))))-----)....))))))))...--- ( -29.20) >DroPer_CAF1 126023 89 + 1 ----------UCACGCUUCAAAGCCGUCACGUCACCAACUAAUUAUCAACAUAAUUAAAAGGUGC-CGUGACUGUACCGGGAGGGUC-----CCCUGACUGAUUA--- ----------....(((....))).((((((.((((...(((((((....)))))))...)))).-)))))).....(((.(((...-----.)))..)))....--- ( -25.70) >consensus __________UUACGCUUCAAAGCCGUCACGUCGCCAACUAAUUAUCAACAUAAUUAAAAGGUGC_CGUGACAGUCCCCGGAGGGUC_____CACUGACUGAUUA___ .................(((.....((((((..(((...(((((((....)))))))...)))...))))))....((....))...............)))...... (-13.86 = -14.12 + 0.25)

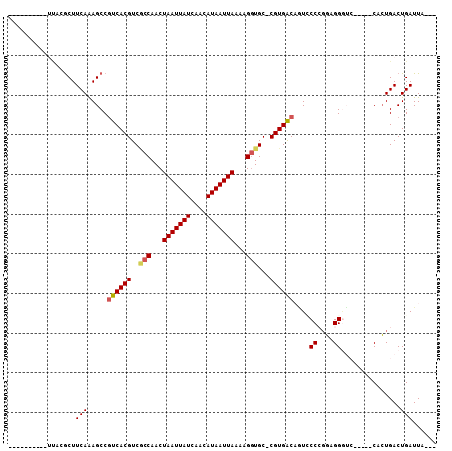
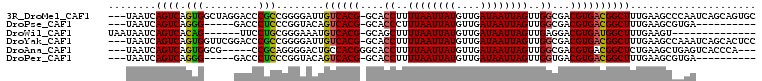
| Location | 14,715,123 – 14,715,227 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -18.29 |
| Energy contribution | -17.65 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14715123 104 - 27905053 ---UAAUCAGUCAGUGGCUAGGACCCGCCGGGGAUUGUCACG-GCACCUUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUUUGAAGCCCAAUCAGCAGUGC ---.....((((..((((........))))..))))((((((-.(.((..((((((((....))))))))..)).).))))))((((....))))............. ( -31.60) >DroPse_CAF1 123589 89 - 1 ---UAAUCAGUCAGGG-----GACCCUCCCGGUACAGUCACG-GCACCCUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUUUGAAGCGUGA---------- ---.....((((.(((-----(....))))......((((((-.(.((..((((((((....))))))))..)).).))))))))))...........---------- ( -26.10) >DroWil_CAF1 151076 87 - 1 UAAUAAUCAGUCACAG------UUCCUGCGGGAAAUGUCACG-GCAGCUUUUAAUUAUGUUGAUAAUUAGUUGAGGACGUGAUGGCUUUGAAGU-------------- ........(((((((.------((((....)))).))(((((-.(..(..((((((((....))))))))..)..).)))))))))).......-------------- ( -20.00) >DroYak_CAF1 135104 104 - 1 ---UAAUCAGUCAGUGGUUCGGACCCGCCGGGGAUUGUCACG-GCACCUUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUUUGAAGCCAAAUCAGCACUCC ---.........((((.(((((.....)))))((((((((((-.(.((..((((((((....))))))))..)).).))))))((((....)))).))))..)))).. ( -33.10) >DroAna_CAF1 134192 97 - 1 ---UAAUCAGUCAGUGGCG-----CCGCAGGGGACUGCCACGGGCACCUUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUCUGAAGCUGAGUCACCCA--- ---...(((((...(((.(-----((((((....))))((((..(.((..((((((((....))))))))..)).).))))..))).)))..)))))........--- ( -35.10) >DroPer_CAF1 126023 89 - 1 ---UAAUCAGUCAGGG-----GACCCUCCCGGUACAGUCACG-GCACCUUUUAAUUAUGUUGAUAAUUAGUUGGUGACGUGACGGCUUUGAAGCGUGA---------- ---.....((((.(((-----(....))))......((((((-.((((..((((((((....))))))))..)))).))))))))))...........---------- ( -30.90) >consensus ___UAAUCAGUCAGUG_____GACCCGCCGGGGACUGUCACG_GCACCUUUUAAUUAUGUUGAUAAUUAGUUGGCGACGUGACGGCUUUGAAGCCUAA__________ ........((((.(((.........)))........((((((....((..((((((((....))))))))..))...))))))))))..................... (-18.29 = -17.65 + -0.64)
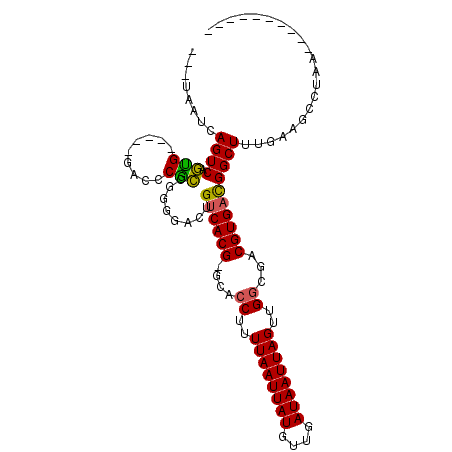
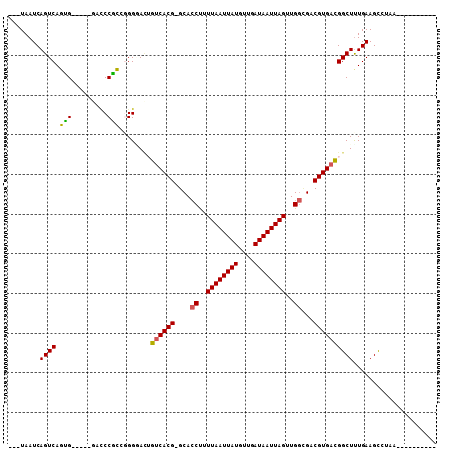
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:12 2006