
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,703,612 – 14,703,717 |
| Length | 105 |
| Max. P | 0.993390 |

| Location | 14,703,612 – 14,703,717 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -13.69 |
| Energy contribution | -13.58 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
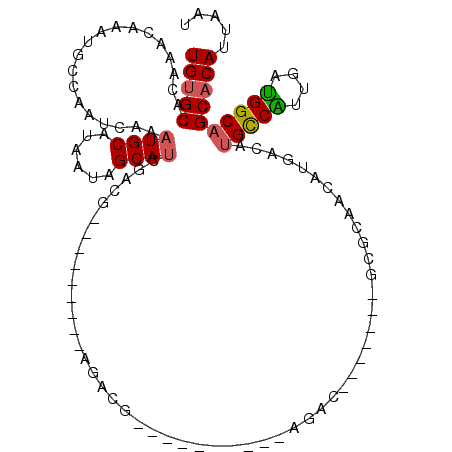
>3R_DroMel_CAF1 14703612 105 + 27905053 UGUGCACAAACAAAUGCCAAUCAAAUGCAUAAUGGCAUCGACGAACAAAACGAGACG----------AGACGAGACGCGCUCAACAUGACAUGUCGCUGAUGGCAGCACAUUAAU (((((........((((.........))))..((.(((((.(((.((...((.(..(----------((.((.....)))))..).))...))))).))))).)))))))..... ( -23.90) >DroPse_CAF1 113590 89 + 1 UGUGCACAAACAAAUGCCAAUCAAAUGCAUAAUAGCAUCGAAG---------CGACG----------CGUC-------AUGGAACAUGACAUGCCAUUGAUGGCAGCACAUUAAU (((((.........(((((.((((..((((....((.(((...---------))).)----------)(((-------(((...))))))))))..))))))))))))))..... ( -27.50) >DroWil_CAF1 138752 87 + 1 UGCGCACAAACAAAUGCCAAUCAAAUGCAUAACAGCAUCGACGAGAGCCAUGA----------------------------GAACAUGACACGUCGUUGACGGCAGCACAUUAAU ((((......)...((((.......(((......)))((((((((.(.((((.----------------------------...)))).).).))))))).)))))))....... ( -20.00) >DroYak_CAF1 123326 108 + 1 UGUGCACAAACAAAUGCCAAUCAAAUGCAUAAUGGCAUCGACGAACAAAACGAUACGAAACGAAACAAGAC-------GCCCAACAUGACAUGCCGCUGAUGGCAGCACAUUAAU (((((.........(((((.(((.((((((...(((((((..........)))).((...)).........-------)))....))).))).....)))))))))))))..... ( -19.00) >DroAna_CAF1 123503 84 + 1 UGUGCGCAAACAAAUGCCAAUCAAAUGCAUAAUAGCACCGACG------------------------ACAU-------GCACAACAUGACAUGCCAUUGAUGGCAGCACAUUAAU ((((((....)...(((((.((((.(((......))).....(------------------------.(((-------(((.....)).)))).).))))))))))))))..... ( -19.90) >DroPer_CAF1 116044 89 + 1 UGUGCACAAACAAAUGCCAAUCAAAUGCAUAAUAGCAUCGAAG---------CGACG----------CGUC-------AUGGAACAUGACAUGCCAUUGAUGGCAGCACAUUAAU (((((.........(((((.((((..((((....((.(((...---------))).)----------)(((-------(((...))))))))))..))))))))))))))..... ( -27.50) >consensus UGUGCACAAACAAAUGCCAAUCAAAUGCAUAAUAGCAUCGACG_________AGACG__________AGAC_______GCGCAACAUGACAUGCCAUUGAUGGCAGCACAUUAAU (((((...................((((......)))).....................................................(((((....))))))))))..... (-13.69 = -13.58 + -0.11)

| Location | 14,703,612 – 14,703,717 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -17.71 |
| Energy contribution | -16.93 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14703612 105 - 27905053 AUUAAUGUGCUGCCAUCAGCGACAUGUCAUGUUGAGCGCGUCUCGUCU----------CGUCUCGUUUUGUUCGUCGAUGCCAUUAUGCAUUUGAUUGGCAUUUGUUUGUGCACA .....((((((((((((((((((....((...((((.(((........----------)))))))...))...))))((((......)))))))).))))).........))))) ( -26.00) >DroPse_CAF1 113590 89 - 1 AUUAAUGUGCUGCCAUCAAUGGCAUGUCAUGUUCCAU-------GACG----------CGUCG---------CUUCGAUGCUAUUAUGCAUUUGAUUGGCAUUUGUUUGUGCACA .....((((((((((((((..((((((((((...)))-------)))(----------(((((---------...))))))....))))..)))).))))).........))))) ( -31.20) >DroWil_CAF1 138752 87 - 1 AUUAAUGUGCUGCCGUCAACGACGUGUCAUGUUC----------------------------UCAUGGCUCUCGUCGAUGCUGUUAUGCAUUUGAUUGGCAUUUGUUUGUGCGCA .....(((((((((((((((((((.((((((...----------------------------.))))))...)))))((((......)))))))).))))).........))))) ( -30.80) >DroYak_CAF1 123326 108 - 1 AUUAAUGUGCUGCCAUCAGCGGCAUGUCAUGUUGGGC-------GUCUUGUUUCGUUUCGUAUCGUUUUGUUCGUCGAUGCCAUUAUGCAUUUGAUUGGCAUUUGUUUGUGCACA .....((((((((((((((..(((((..(((..((((-------(........))))).((((((..........))))))))))))))..)))).))))).........))))) ( -26.30) >DroAna_CAF1 123503 84 - 1 AUUAAUGUGCUGCCAUCAAUGGCAUGUCAUGUUGUGC-------AUGU------------------------CGUCGGUGCUAUUAUGCAUUUGAUUGGCAUUUGUUUGCGCACA .....((((((((((((((((((((((........))-------))))------------------------))).(((((......)))))))).)))))...(....)))))) ( -26.40) >DroPer_CAF1 116044 89 - 1 AUUAAUGUGCUGCCAUCAAUGGCAUGUCAUGUUCCAU-------GACG----------CGUCG---------CUUCGAUGCUAUUAUGCAUUUGAUUGGCAUUUGUUUGUGCACA .....((((((((((((((..((((((((((...)))-------)))(----------(((((---------...))))))....))))..)))).))))).........))))) ( -31.20) >consensus AUUAAUGUGCUGCCAUCAACGGCAUGUCAUGUUCCGC_______GUCU__________CGUCU_________CGUCGAUGCUAUUAUGCAUUUGAUUGGCAUUUGUUUGUGCACA .....(((((((((((((..(((((...)))))...........................................(((((......)))))))).))))).........))))) (-17.71 = -16.93 + -0.78)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:06 2006