
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,694,905 – 14,695,042 |
| Length | 137 |
| Max. P | 0.980475 |

| Location | 14,694,905 – 14,695,002 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -40.57 |
| Consensus MFE | -24.27 |
| Energy contribution | -25.58 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -5.88 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14694905 97 + 27905053 CUAUUUUAAUUAAUUCCAGGCAACCGCAAGGCAAUCAACUUGCGGUCAGGCGA--------UGGAGUUUUCCACCAUCAAAU-----AUUGAUGGGG----------AAAACUCUGUCUU ...................((.((((((((........))))))))...))((--------(((((((((((.((((((...-----..))))))))----------))))))))))).. ( -43.40) >DroSec_CAF1 113904 97 + 1 CUAUUUUAAUUAAUUCCAGGCAACCGCAAGGCAAUCAACUUGCGGUCAGGCGA--------UGGAGUUUUCCACCAUCAAAU-----AUUGAUGGGG----------AAAACUCUGUCUU ...................((.((((((((........))))))))...))((--------(((((((((((.((((((...-----..))))))))----------))))))))))).. ( -43.40) >DroSim_CAF1 118184 97 + 1 CUAUUUUAAUUAAUUCCAGGCAACCGCAAGGCAAUCAACUUGCGGUCAGGCGA--------UGGAGUUUUCCACCAUCAAAU-----AUUGAUGGGG----------AAAACUCUGUCUU ...................((.((((((((........))))))))...))((--------(((((((((((.((((((...-----..))))))))----------))))))))))).. ( -43.40) >DroEre_CAF1 109230 97 + 1 CUAUUUUAAUUAAUUCCAGGCAACCGCAAGGCAAUCAACUUGCGGUCAGGCGA--------UGGAGUUUUCCACCAUCAAAU-----AUUGAUGGGG----------AAAAUUCUGUCUU ...................((.((((((((........))))))))...))((--------(((((((((((.((((((...-----..))))))))----------))))))))))).. ( -41.10) >DroYak_CAF1 114833 97 + 1 CUAUUUUAAUUAAUUCCAGGCAACCGCAAGGCAAUCAACUUGCGGUCAGGCGA--------UGAAGUUUUCCACCAUCAAAU-----AUUGAUGGGG----------AAAACUCUGUCUU ...................((.((((((((........))))))))...))((--------(..((((((((.((((((...-----..))))))))----------))))))..))).. ( -38.40) >DroPer_CAF1 108117 119 + 1 UUUUUUUAAUUAAUUUCUGGCCACCGCAAGGCAGCAAACCUGUGGUCGGGCCAACCCCUGGCAGACCCCUGGAUCAUAGAAUGGAGAAUGGAUGGAGGAGCCCAGGCAAAACCCUCCCC- .(((((((......(((((((..((....))..))...((.(.((((..((((.....)))).)))).).))....)))))))))))).....(((((.((....)).....)))))..- ( -33.70) >consensus CUAUUUUAAUUAAUUCCAGGCAACCGCAAGGCAAUCAACUUGCGGUCAGGCGA________UGGAGUUUUCCACCAUCAAAU_____AUUGAUGGGG__________AAAACUCUGUCUU ...................((.((((((((........))))))))...))...........((((((((((.(((((((........)))))))))..........))))))))..... (-24.27 = -25.58 + 1.31)



| Location | 14,694,905 – 14,695,002 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -22.85 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.22 |
| Mean z-score | -4.07 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14694905 97 - 27905053 AAGACAGAGUUUU----------CCCCAUCAAU-----AUUUGAUGGUGGAAAACUCCA--------UCGCCUGACCGCAAGUUGAUUGCCUUGCGGUUGCCUGGAAUUAAUUAAAAUAG ..((..(((((((----------((((((((..-----...)))))).)))))))))..--------)).((.(((((((((........)))))))))....))............... ( -37.30) >DroSec_CAF1 113904 97 - 1 AAGACAGAGUUUU----------CCCCAUCAAU-----AUUUGAUGGUGGAAAACUCCA--------UCGCCUGACCGCAAGUUGAUUGCCUUGCGGUUGCCUGGAAUUAAUUAAAAUAG ..((..(((((((----------((((((((..-----...)))))).)))))))))..--------)).((.(((((((((........)))))))))....))............... ( -37.30) >DroSim_CAF1 118184 97 - 1 AAGACAGAGUUUU----------CCCCAUCAAU-----AUUUGAUGGUGGAAAACUCCA--------UCGCCUGACCGCAAGUUGAUUGCCUUGCGGUUGCCUGGAAUUAAUUAAAAUAG ..((..(((((((----------((((((((..-----...)))))).)))))))))..--------)).((.(((((((((........)))))))))....))............... ( -37.30) >DroEre_CAF1 109230 97 - 1 AAGACAGAAUUUU----------CCCCAUCAAU-----AUUUGAUGGUGGAAAACUCCA--------UCGCCUGACCGCAAGUUGAUUGCCUUGCGGUUGCCUGGAAUUAAUUAAAAUAG .........((((----------((((((((..-----...)))))).)))))).((((--------..((..(((((((((........))))))))))).)))).............. ( -32.60) >DroYak_CAF1 114833 97 - 1 AAGACAGAGUUUU----------CCCCAUCAAU-----AUUUGAUGGUGGAAAACUUCA--------UCGCCUGACCGCAAGUUGAUUGCCUUGCGGUUGCCUGGAAUUAAUUAAAAUAG ..((..(((((((----------((((((((..-----...)))))).)))))))))..--------)).((.(((((((((........)))))))))....))............... ( -35.80) >DroPer_CAF1 108117 119 - 1 -GGGGAGGGUUUUGCCUGGGCUCCUCCAUCCAUUCUCCAUUCUAUGAUCCAGGGGUCUGCCAGGGGUUGGCCCGACCACAGGUUUGCUGCCUUGCGGUGGCCAGAAAUUAAUUAAAAAAA -(((((((((((.....)))).)))))..))........((((..(((((..((.....))..)))))((((..(((.(((((.....))).)).))))))))))).............. ( -36.80) >consensus AAGACAGAGUUUU__________CCCCAUCAAU_____AUUUGAUGGUGGAAAACUCCA________UCGCCUGACCGCAAGUUGAUUGCCUUGCGGUUGCCUGGAAUUAAUUAAAAUAG ......(((((((..........(((((((((........))))))).)))))))))............((..(((((((((........)))))))))))................... (-22.85 = -23.52 + 0.67)

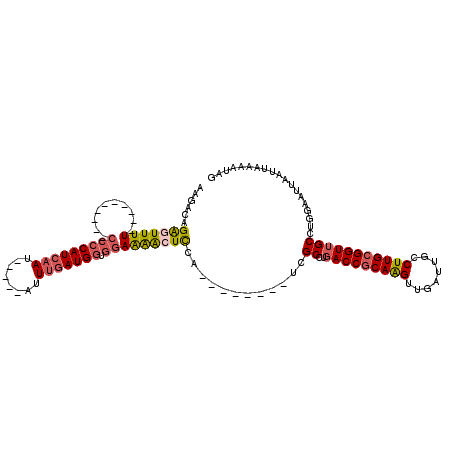
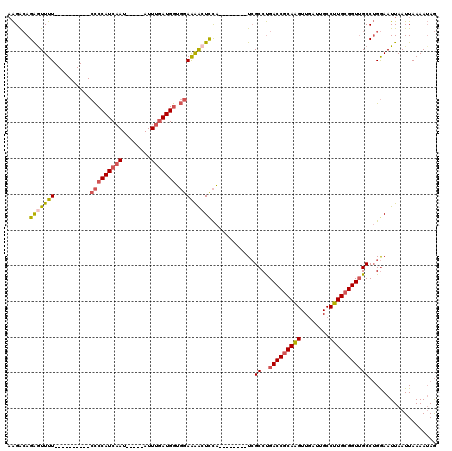
| Location | 14,694,945 – 14,695,042 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.91 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -17.96 |
| Energy contribution | -19.68 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14694945 97 + 27905053 UGCGGUCAGGCGA--------UGGAGUUUUCCACCAUCAAAU-----AUUGAUGGGG----------AAAACUCUGUCUUCGGCAGCGACCAAGCGUAAGCAGCUCAAUUUCAAGCAGCU .((((((..((((--------(((((((((((.((((((...-----..))))))))----------)))))))))))....))...))))..((....)).(((........))).)). ( -41.80) >DroSec_CAF1 113944 97 + 1 UGCGGUCAGGCGA--------UGGAGUUUUCCACCAUCAAAU-----AUUGAUGGGG----------AAAACUCUGUCUUCGGCAGCGACCAAGCGUAAGCAGCUCAAUUUCAAGUAGCU .((((((..((((--------(((((((((((.((((((...-----..))))))))----------)))))))))))....))...))))..((....)).))................ ( -40.20) >DroSim_CAF1 118224 97 + 1 UGCGGUCAGGCGA--------UGGAGUUUUCCACCAUCAAAU-----AUUGAUGGGG----------AAAACUCUGUCUUCGGCAGCGACCAAGCGUAAGCAGCUCAAUUUCAAGCAGCU .((((((..((((--------(((((((((((.((((((...-----..))))))))----------)))))))))))....))...))))..((....)).(((........))).)). ( -41.80) >DroEre_CAF1 109270 97 + 1 UGCGGUCAGGCGA--------UGGAGUUUUCCACCAUCAAAU-----AUUGAUGGGG----------AAAAUUCUGUCUUCGGCAGCGACCAAGCGUAAGCAGCUCAAUUUCAAGCAGCU .((((((..((((--------(((((((((((.((((((...-----..))))))))----------)))))))))))....))...))))..((....)).(((........))).)). ( -39.50) >DroYak_CAF1 114873 97 + 1 UGCGGUCAGGCGA--------UGAAGUUUUCCACCAUCAAAU-----AUUGAUGGGG----------AAAACUCUGUCUUCGGCAGCGACCAAGCGUAAGCAGCUCAAUUUCAAGCAGCU .((((((..((((--------(..((((((((.((((((...-----..))))))))----------))))))..)))....))...))))..((....)).(((........))).)). ( -36.80) >DroPer_CAF1 108157 114 + 1 UGUGGUCGGGCCAACCCCUGGCAGACCCCUGGAUCAUAGAAUGGAGAAUGGAUGGAGGAGCCCAGGCAAAACCCUCCCC----AAACG--UAAGCGUAAGCAGCUCAAUUUCGAGAAGCU ...((((..((((.....)))).))))...........(((..(((.(((..(((.((((....((.....))))))))----)..))--)..((....))..)))...)))........ ( -32.20) >consensus UGCGGUCAGGCGA________UGGAGUUUUCCACCAUCAAAU_____AUUGAUGGGG__________AAAACUCUGUCUUCGGCAGCGACCAAGCGUAAGCAGCUCAAUUUCAAGCAGCU .((((((...............((((((((((.(((((((........)))))))))..........))))))))((.....))...))))..))...(((.(((........))).))) (-17.96 = -19.68 + 1.73)
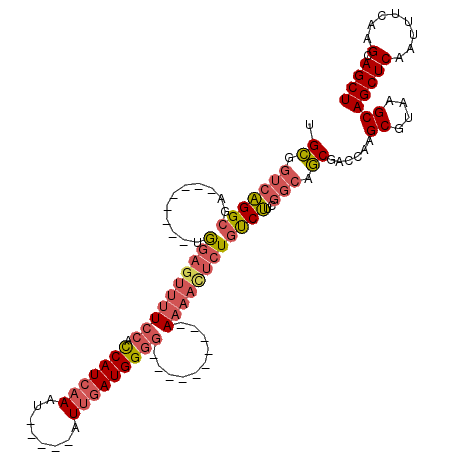
| Location | 14,694,945 – 14,695,042 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.91 |
| Mean single sequence MFE | -34.59 |
| Consensus MFE | -19.48 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14694945 97 - 27905053 AGCUGCUUGAAAUUGAGCUGCUUACGCUUGGUCGCUGCCGAAGACAGAGUUUU----------CCCCAUCAAU-----AUUUGAUGGUGGAAAACUCCA--------UCGCCUGACCGCA (((.((((......)))).)))...((..(((((..((....((..(((((((----------((((((((..-----...)))))).)))))))))..--------)))).))))))). ( -36.50) >DroSec_CAF1 113944 97 - 1 AGCUACUUGAAAUUGAGCUGCUUACGCUUGGUCGCUGCCGAAGACAGAGUUUU----------CCCCAUCAAU-----AUUUGAUGGUGGAAAACUCCA--------UCGCCUGACCGCA .((...........((((.......))))(((((..((....((..(((((((----------((((((((..-----...)))))).)))))))))..--------)))).))))))). ( -33.70) >DroSim_CAF1 118224 97 - 1 AGCUGCUUGAAAUUGAGCUGCUUACGCUUGGUCGCUGCCGAAGACAGAGUUUU----------CCCCAUCAAU-----AUUUGAUGGUGGAAAACUCCA--------UCGCCUGACCGCA (((.((((......)))).)))...((..(((((..((....((..(((((((----------((((((((..-----...)))))).)))))))))..--------)))).))))))). ( -36.50) >DroEre_CAF1 109270 97 - 1 AGCUGCUUGAAAUUGAGCUGCUUACGCUUGGUCGCUGCCGAAGACAGAAUUUU----------CCCCAUCAAU-----AUUUGAUGGUGGAAAACUCCA--------UCGCCUGACCGCA (((.((((......)))).)))...((..(((((..((....((..((.((((----------((((((((..-----...)))))).)))))).))..--------)))).))))))). ( -30.50) >DroYak_CAF1 114873 97 - 1 AGCUGCUUGAAAUUGAGCUGCUUACGCUUGGUCGCUGCCGAAGACAGAGUUUU----------CCCCAUCAAU-----AUUUGAUGGUGGAAAACUUCA--------UCGCCUGACCGCA (((.((((......)))).)))...((..(((((..((.((.((...((((((----------((((((((..-----...)))))).)))))))))).--------)))).))))))). ( -35.10) >DroPer_CAF1 108157 114 - 1 AGCUUCUCGAAAUUGAGCUGCUUACGCUUA--CGUUU----GGGGAGGGUUUUGCCUGGGCUCCUCCAUCCAUUCUCCAUUCUAUGAUCCAGGGGUCUGCCAGGGGUUGGCCCGACCACA ..(((((((((..(((((.......)))))--..)))----))))))((((......(((((.((((...((..((((.............))))..))...))))..)))))))))... ( -35.22) >consensus AGCUGCUUGAAAUUGAGCUGCUUACGCUUGGUCGCUGCCGAAGACAGAGUUUU__________CCCCAUCAAU_____AUUUGAUGGUGGAAAACUCCA________UCGCCUGACCGCA (((.((((......)))).)))...((..(((((((.....))...(((((((..........(((((((((........))))))).)))))))))...............))))))). (-19.48 = -20.40 + 0.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:01 2006