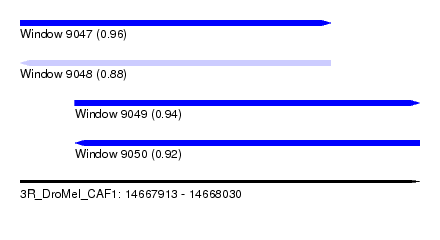
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,667,913 – 14,668,030 |
| Length | 117 |
| Max. P | 0.957301 |

| Location | 14,667,913 – 14,668,004 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -17.27 |
| Energy contribution | -17.05 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14667913 91 + 27905053 UGGGAGAAAA-----AAAAAAAAAAAACGGUGGGCGCCAGUUAGUGGUGCGUGCGCCAUAAGCUAAAAUGUCAAUAUUUGACUAUGCUACGCAUAA ..........-----.............(((....)))((((.((((((....)))))).)))).....((((.....))))(((((...))))). ( -20.10) >DroPse_CAF1 80640 77 + 1 UGU-----GGCUGG--------------GGCAGGCGCCAGUUAGUGGUGCGUGCGCCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCCACGCAUAA .((-----(((.((--------------.(((.((((((.....)))))).))).))(((.((..(((((....))))))).))))))))...... ( -30.30) >DroSim_CAF1 90281 96 + 1 UGCGAGGGAGCUGGUGAAUAAAAAAAGCGUUGGGCGCCAGUUAGUGGUGCGUGCGCCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCUACGCAUAA ((((....(((.((..((((......(((.....))).((((.((((((....)))))).))))..........))))..))...))).))))... ( -30.70) >DroEre_CAF1 81937 88 + 1 --CGAGGGAGCUGG------GAAAAAAUGGUGGGCGCCAGUUAGUGGUGCGUGCGCCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCUCCGCAUAA --....((((((((------(...(((((...((((..((((.((((((....)))))).))))....))))..))))).)))).)))))...... ( -31.50) >DroAna_CAF1 79441 85 + 1 ------GGCACUGGG-----GAAAGAAUGGCGGGCGCCAGUUAGUGGUGCGUGCCCCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCCACGCAUAA ------((((.((((-----.(((....((((.((((((.....)))))).)))).(((........)))......))).))))))))........ ( -29.20) >DroPer_CAF1 82552 77 + 1 UGU-----GGCUGG--------------GGCAGGCGCCAGUUAGUGGUGCGUGCGCCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCCACGCAUAA .((-----(((.((--------------.(((.((((((.....)))))).))).))(((.((..(((((....))))))).))))))))...... ( -30.30) >consensus UGC___GGAGCUGG_______AAAAAA_GGCGGGCGCCAGUUAGUGGUGCGUGCGCCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCCACGCAUAA ............................(((((((...((((.((((((....)))))).)))).(((((....)))))))))..)))........ (-17.27 = -17.05 + -0.22)



| Location | 14,667,913 – 14,668,004 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -15.05 |
| Energy contribution | -15.55 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14667913 91 - 27905053 UUAUGCGUAGCAUAGUCAAAUAUUGACAUUUUAGCUUAUGGCGCACGCACCACUAACUGGCGCCCACCGUUUUUUUUUUUU-----UUUUCUCCCA ...(((((.((...((((.....))))......((.....)))))))))(((.....))).....................-----.......... ( -15.20) >DroPse_CAF1 80640 77 - 1 UUAUGCGUGGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGGCGCACGCACCACUAACUGGCGCCUGCC--------------CCAGCC-----ACA ......((((((((((((((.........))).))))))((.(((.((.(((.....))).)).))).--------------)).)))-----)). ( -26.80) >DroSim_CAF1 90281 96 - 1 UUAUGCGUAGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGGCGCACGCACCACUAACUGGCGCCCAACGCUUUUUUUAUUCACCAGCUCCCUCGCA ...((((.((((((((((((.........))).))))))((((...((.(((.....))).))....))))..............)))....)))) ( -19.80) >DroEre_CAF1 81937 88 - 1 UUAUGCGGAGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGGCGCACGCACCACUAACUGGCGCCCACCAUUUUUUC------CCAGCUCCCUCG-- ......((((((((((((((.........))).))))))(((((.((..........)))))))............------...)))))....-- ( -21.30) >DroAna_CAF1 79441 85 - 1 UUAUGCGUGGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGGGGCACGCACCACUAACUGGCGCCCGCCAUUCUUUC-----CCCAGUGCC------ ....((((((((((((((((.........))).))))))).(((..((.(((.....))).))..)))........-----.))).))).------ ( -22.00) >DroPer_CAF1 82552 77 - 1 UUAUGCGUGGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGGCGCACGCACCACUAACUGGCGCCUGCC--------------CCAGCC-----ACA ......((((((((((((((.........))).))))))((.(((.((.(((.....))).)).))).--------------)).)))-----)). ( -26.80) >consensus UUAUGCGUAGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGGCGCACGCACCACUAACUGGCGCCCACC_UUUUUU_______CCAGCUCC___ACA ...(((((..((((((((((.........))).)))))))))))).((.(((.....))).))................................. (-15.05 = -15.55 + 0.50)



| Location | 14,667,929 – 14,668,030 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.73 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -23.09 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14667929 101 + 27905053 AAAAAACGGUGGGCGCCAGUUAGUGGUGCGUGCGCCAUAAGCUAAAAUGUCAAUAUUUGACUAUGCUACGCAUAAAA--UGCCAAGGCAUAUGCCUUGUGGUG--------- ........(((.((((((.....)))))).)))(((((((((......((((.....))))((((((..((((...)--)))...)))))).).)))))))).--------- ( -32.40) >DroVir_CAF1 96580 112 + 1 CCAAAACGGGAAGCGUCAGUUAGUGGCGCGUGCUCCAUAAGCCCAAAUGUCAAUAUUUGCCUAUGCCAGGCAUUAAACGCGCCAAAGCAUGUGCCUCAUAGUAUGUGUUUGG ((((((((.((.(((.(((((..(((((((((((.....)))...............(((((.....)))))....)))))))).))).))))).))......))).))))) ( -31.80) >DroPse_CAF1 80649 96 + 1 -------GGCAGGCGCCAGUUAGUGGUGCGUGCGCCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCCACGCAUAAAA--UGCCAAGGCAUAUGCCUUCUCGGAGG------- -------(((((((...((((.((((((....)))))).)))).(((((....))))))))((((((..((((...)--)))...))))))))))..........------- ( -31.30) >DroWil_CAF1 98302 94 + 1 UGUAGGCAGUUGGCGCCAGUUAGUGGUGCGUGCGGCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCCACGCAUAAAA--UGCCAUAGCAUAUGCCG---------------- ....((((....((((((.....))))))(((.((((((.((..(((((....))))))).)))))).))).....(--(((....)))).)))).---------------- ( -32.90) >DroAna_CAF1 79451 100 + 1 AAAGAAUGGCGGGCGCCAGUUAGUGGUGCGUGCCCCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCCACGCAUAAAG--UGCCAAGGCAUAUGCCUUGUGGU---------- .......((((.((((((.....)))))).))))(((((((...(((((....)))))((.((((((..((((...)--)))...)))))).))))))))).---------- ( -36.80) >DroPer_CAF1 82561 97 + 1 -------GGCAGGCGCCAGUUAGUGGUGCGUGCGCCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCCACGCAUAAAA--UGCCAAGGCAUAUGCCUUCUCGGAGGC------ -------((((((((..((((.((((((....)))))).))))....))))......))))((((((..((((...)--)))...)))))).(((((....)))))------ ( -32.80) >consensus __AAAACGGCAGGCGCCAGUUAGUGGUGCGUGCGCCAUAAGCUAAAAUGUCAAUAUUUGCCUAUGCCACGCAUAAAA__UGCCAAGGCAUAUGCCUUCUCGUA_________ .......(((..((((((.....))))))(((((.((((.((..(((((....))))))).))))...))))).......)))(((((....)))))............... (-23.09 = -23.48 + 0.39)


| Location | 14,667,929 – 14,668,030 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.73 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -22.31 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14667929 101 - 27905053 ---------CACCACAAGGCAUAUGCCUUGGCA--UUUUAUGCGUAGCAUAGUCAAAUAUUGACAUUUUAGCUUAUGGCGCACGCACCACUAACUGGCGCCCACCGUUUUUU ---------.....((((((....))))))((.--.....(((((.((...((((.....))))......((.....)))))))))(((.....))).))............ ( -26.60) >DroVir_CAF1 96580 112 - 1 CCAAACACAUACUAUGAGGCACAUGCUUUGGCGCGUUUAAUGCCUGGCAUAGGCAAAUAUUGACAUUUGGGCUUAUGGAGCACGCGCCACUAACUGACGCUUCCCGUUUUGG (((((.((.......(((((....)))))(((((((....((((((...))))))...............((((...))))))))))).................))))))) ( -30.60) >DroPse_CAF1 80649 96 - 1 -------CCUCCGAGAAGGCAUAUGCCUUGGCA--UUUUAUGCGUGGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGGCGCACGCACCACUAACUGGCGCCUGCC------- -------........(((((....)))))(((.--.....(((((((((((((((((.........))).)))))).)).))))))(((.....))).)))....------- ( -29.70) >DroWil_CAF1 98302 94 - 1 ----------------CGGCAUAUGCUAUGGCA--UUUUAUGCGUGGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGCCGCACGCACCACUAACUGGCGCCAACUGCCUACA ----------------.((((.((((....)))--).....(.((((((((((((((.........))).))))))))))).)((.(((.....))).))....)))).... ( -31.10) >DroAna_CAF1 79451 100 - 1 ----------ACCACAAGGCAUAUGCCUUGGCA--CUUUAUGCGUGGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGGGGCACGCACCACUAACUGGCGCCCGCCAUUCUUU ----------.(((((((((....))))).(((--.....)))))))((((((((((.........))).))))))).(((..((.(((.....))).))..)))....... ( -31.30) >DroPer_CAF1 82561 97 - 1 ------GCCUCCGAGAAGGCAUAUGCCUUGGCA--UUUUAUGCGUGGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGGCGCACGCACCACUAACUGGCGCCUGCC------- ------((.......(((((....)))))(((.--.....(((((((((((((((((.........))).)))))).)).))))))(((.....))).))).)).------- ( -30.10) >consensus _________UACCACAAGGCAUAUGCCUUGGCA__UUUUAUGCGUGGCAUAGGCAAAUAUUGACAUUUUAGCUUAUGGCGCACGCACCACUAACUGGCGCCUGCCGUUUU__ ...............(((((....)))))(((........((((((.(((((((((((......))))..)))))))...))))))(((.....))).)))........... (-22.31 = -22.70 + 0.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:52 2006