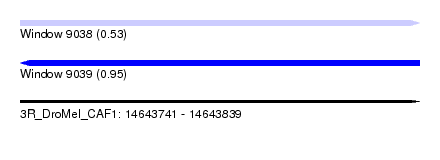
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,643,741 – 14,643,839 |
| Length | 98 |
| Max. P | 0.954210 |

| Location | 14,643,741 – 14,643,839 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 67.42 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -2.78 |
| Energy contribution | -3.42 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.11 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14643741 98 + 27905053 UUACGGCACCCAAGCACCACACACACACACGCCACG---CAUG--UGAGUGUGUGCGUGAGUGCAAUUUCAUUAAUGGCUAAUUAAUUGCAACUUUAUUUCGC .....(((.....(((((((.(((((((.(((....---...)--)).))))))).))).))))......((((((.....)))))))))............. ( -28.90) >DroVir_CAF1 66538 95 + 1 UUACGCAAUGCAAGCUGGAAAUUAAGC-A--C-----UGUAUGUCGGAUUGUGCCUGUGUGUUCUGUUUUAAUAUGAAACACUUAAUUGCAGCUUUAUUUCGU ..(((.((((.((((((..((((((((-(--(-----.(((((......)))))..))).....((((((.....))))))))))))).)))))))))).))) ( -22.70) >DroSec_CAF1 58927 101 + 1 UUACGCCACCCAAGCACCACACACACACACACCAUGAUGUGUG--UGCGUGUGUGCGUGAGUGCAAUUUCAUUAAUGGCUAAUUAAUUGCAACUUUAUUUCGC ....((........((((((((((.(((((((......)))))--)).))))))).)))..(((((((..((((.....)))).)))))))..........)) ( -32.90) >DroSim_CAF1 65899 93 + 1 UUACGCCACCCAAGCACCACACACACACACACCAUGAUGUGUG--U--------GCGUGAGUGCAAUUUCAUUAAUGGCUAAUUAAUUGCAACUUUAUUUCGC ....(((......(((((((.(((((((((.....).))))))--)--------).))).)))).(((.....))))))........................ ( -26.40) >DroEre_CAF1 57488 84 + 1 UUACGCCACCCAAGCACCG------------CCAUG-----UG--UGAGUGUGUGCGUGAGUGCAAUUUCAUUAAUGGCUAAUUAAUUGCAACUUUAUUUCGC (((((((((.(..((((..------------....)-----))--)..).))).)))))).(((((((..((((.....)))).)))))))............ ( -21.10) >DroMoj_CAF1 66860 84 + 1 UUACGGCGCACAAGCUGCAAAUGAACG-A--G-----C----------AUGUGUGCGUGUGUUCCGCUUUGAUAUGA-GCAAUUAAUUGCAGCUUUAUUUCGU ..((((.....(((((((((.(((..(-(--(-----(----------(..(....)..))))).((((......))-))..))).)))))))))....)))) ( -25.70) >consensus UUACGCCACCCAAGCACCACACACACA_A__CCAUG_U_U_UG__UGAGUGUGUGCGUGAGUGCAAUUUCAUUAAUGGCUAAUUAAUUGCAACUUUAUUUCGC ......................................................(((.((((((((((................)))))).)))).....))) ( -2.78 = -3.42 + 0.64)


| Location | 14,643,741 – 14,643,839 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 67.42 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -4.61 |
| Energy contribution | -5.45 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.15 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
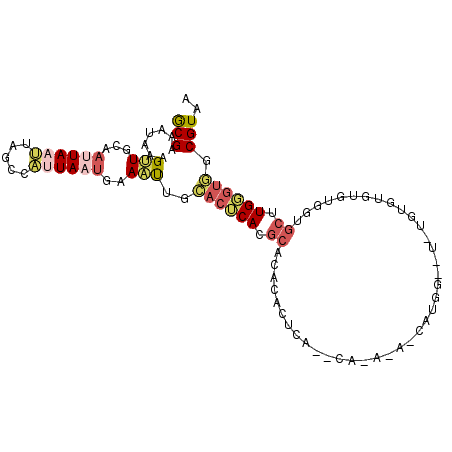
>3R_DroMel_CAF1 14643741 98 - 27905053 GCGAAAUAAAGUUGCAAUUAAUUAGCCAUUAAUGAAAUUGCACUCACGCACACACUCA--CAUG---CGUGGCGUGUGUGUGUGUGGUGCUUGGGUGCCGUAA (((......((((...((((((.....))))))..))))(((((((.((((((((.((--((((---((...)))))))).)))).)))).)))))))))).. ( -36.20) >DroVir_CAF1 66538 95 - 1 ACGAAAUAAAGCUGCAAUUAAGUGUUUCAUAUUAAAACAGAACACACAGGCACAAUCCGACAUACA-----G--U-GCUUAAUUUCCAGCUUGCAUUGCGUAA (((.(((.((((((.((((((((((((............))))))....((((.............-----)--)-))))))))..))))))..))).))).. ( -20.12) >DroSec_CAF1 58927 101 - 1 GCGAAAUAAAGUUGCAAUUAAUUAGCCAUUAAUGAAAUUGCACUCACGCACACACGCA--CACACAUCAUGGUGUGUGUGUGUGUGGUGCUUGGGUGGCGUAA ((...........((((((.(((((...)))))..))))))(((((.(((((((((((--(((((((....)))))))))))))).)))).))))).)).... ( -43.70) >DroSim_CAF1 65899 93 - 1 GCGAAAUAAAGUUGCAAUUAAUUAGCCAUUAAUGAAAUUGCACUCACGC--------A--CACACAUCAUGGUGUGUGUGUGUGUGGUGCUUGGGUGGCGUAA ((((.......)))).........((((((.........((((((((((--------(--(((((((....))))))))))))).)))))...)))))).... ( -38.40) >DroEre_CAF1 57488 84 - 1 GCGAAAUAAAGUUGCAAUUAAUUAGCCAUUAAUGAAAUUGCACUCACGCACACACUCA--CA-----CAUGG------------CGGUGCUUGGGUGGCGUAA ((...........((((((.(((((...)))))..))))))(((((.((((...(...--..-----...).------------..)))).))))).)).... ( -19.10) >DroMoj_CAF1 66860 84 - 1 ACGAAAUAAAGCUGCAAUUAAUUGC-UCAUAUCAAAGCGGAACACACGCACACAU----------G-----C--U-CGUUCAUUUGCAGCUUGUGCGCCGUAA (((.....(((((((((.....(((-(........))))((((....(((....)----------)-----)--.-.))))..)))))))))......))).. ( -21.00) >consensus GCGAAAUAAAGUUGCAAUUAAUUAGCCAUUAAUGAAAUUGCACUCACGCACACACUCA__CA_A_A_CAUGG__U_UGUGUGUGUGGUGCUUGGGUGGCGUAA (((.......(((...((((((.....))))))..)))..((((((.((.......................................)).)))))).))).. ( -4.61 = -5.45 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:42 2006