| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,062,510 – 2,062,611 |
| Length | 101 |
| Max. P | 0.763236 |

| Location | 2,062,510 – 2,062,611 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.31 |
| Mean single sequence MFE | -12.22 |
| Consensus MFE | -9.07 |
| Energy contribution | -8.68 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
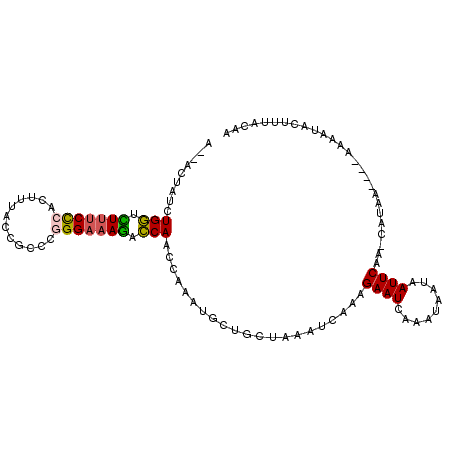
>3R_DroMel_CAF1 2062510 101 + 27905053 G--ACUAUCUGGACUUUCUCACUUUACCGCCCGGGAAAGACCAACCGAAUGCUGCUAAAUCAAAGAAUCAAAUAAUAAUUCAAACAUAA----AAAAUACUUUACCA .--......(((.(((((((............))))))).)))...((((...........................))))........----.............. ( -9.93) >DroPse_CAF1 182236 106 + 1 AAAUCUAUCUGAUUUUUCCAACUUUACCGCAAAGGAAAACUCAAUCAAAUGCUGAUAAAUCAAAGAAUCAAAUAAUAAUUCAA-CAUAAAAGAAAAAUCCUAUACAC ..........((((((((..........(((...((........))...)))............((((.........))))..-.......))))))))........ ( -9.30) >DroSim_CAF1 211525 101 + 1 A--ACUAUCUGGUCUUUCCCACUUUACCGCCCGGGAAAGACCAACCAAAUGCUGCUAAAUCAAAGAAUCAAAUAAUAAUUCAAACAUGA----AAAAUACUUUACAA .--......(((((((((((............)))))))))))...................................((((....)))----)............. ( -18.80) >DroEre_CAF1 206343 93 + 1 A--ACUAUCUGGGUUUCCCCACUUUACCGCCCGGGAAAGACCAACCAAAUGCUGCUAAAUCAAAGAAUCAAAUAAUAAUUCGA------------AAUACUUUACAA .--......(((.((((((.............))))))..))).....................((((.........))))..------------............ ( -9.22) >DroAna_CAF1 182896 105 + 1 A--UCUAUCUGGGCUCUCCCACCUUACCGCCCGGGAAAGCCCAACCAAAUGCUGCUAAAUCAAAGAAUCAAAUAAUAAUUCAAACAUAAACAAAAAAUGCCUAACUA .--......((((((.((((............)))).)))))).....................((((.........)))).......................... ( -16.80) >DroPer_CAF1 184139 106 + 1 AAAUCUAUCUGAUUUUUCCAACUUUACCGCAAAGGAAAACUCAAUCAAAUGCUGAUAAAUCAAAGAAUCAAAUAAUAAUUCAA-CAUAAAAGAAAAAUCCUAUACAC ..........((((((((..........(((...((........))...)))............((((.........))))..-.......))))))))........ ( -9.30) >consensus A__ACUAUCUGGUCUUUCCCACUUUACCGCCCGGGAAAGACCAACCAAAUGCUGCUAAAUCAAAGAAUCAAAUAAUAAUUCAA_CAUAA____AAAAUACUUUACAA .........(((.(((((((............))))))).))).....................((((.........)))).......................... ( -9.07 = -8.68 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:58 2006