| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,622,689 – 14,622,779 |
| Length | 90 |
| Max. P | 0.850943 |

| Location | 14,622,689 – 14,622,779 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.35 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
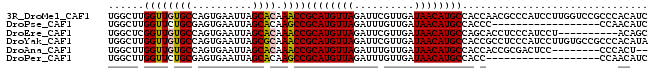
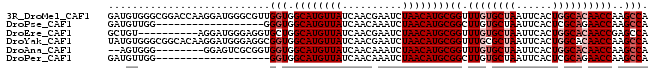
>3R_DroMel_CAF1 14622689 90 + 27905053 UGGCUUGGUUGUGCCAGUGAAUUAGCACAAACCGCAUGUUAGAUUCGUUGAUAACAUGCCACCAACGCCCAUCCUUGGUCCGCCCACAUC .(((((((((((((..........)))))....((((((((..........)))))))).))))).)))......(((.....))).... ( -24.20) >DroPse_CAF1 35650 72 + 1 UGGCUUGGUUCUGCGAGUGAAUUAGCACAAGCCGCAUGUUAGAUUUGUUGAUAACAUGCCACCC------------------CCAACAUC .((((((....(((..........)))))))))((((((((..........)))))))).....------------------........ ( -19.40) >DroEre_CAF1 36275 80 + 1 UGGCUCGGUUGUGCCAGUGAAUUAGCACAAACCGCAUGUUAGAUUCGUUGAUAACAUGCCAGCACCUCCCAUCCU----------ACAGC .((...((..((((..(((......))).....((((((((..........))))))))..))))..))...)).----------..... ( -20.60) >DroYak_CAF1 38091 90 + 1 UGGCUUGGUUGUGCCAGUGAAUUAGCGCAAACCGCAUGUUAGAUUCGUUGAUAACAUGCCACCGCCUCCCAUCCUUGUGCCGCCCACAUA .(((..((((((((..........)))).))))((((((((..........))))))))....))).........((((.....)))).. ( -23.60) >DroAna_CAF1 35122 80 + 1 UGGCUUGGUUGUGCCAGUGAAUUAGCACAAACCGCAUGUUAGAUUUGUUGAUAACAUGCCACCACCGCGACUCC--------CCCACU-- (((...(((((((...(((......))).....((((((((..........))))))))......)))))))..--------.)))..-- ( -20.60) >DroPer_CAF1 38304 71 + 1 UGGCUUGGUUCUGCGAGUGAAUUAGCACAAGCCGCAUGUUAGAUUUGUUGAUAACAUGCCACC-------------------CCAACAUC .((((((....(((..........)))))))))((((((((..........))))))))....-------------------........ ( -19.40) >consensus UGGCUUGGUUGUGCCAGUGAAUUAGCACAAACCGCAUGUUAGAUUCGUUGAUAACAUGCCACCACC_CCCAUCC________CCCACAUC ......((((((((..........)))).))))((((((((..........))))))))............................... (-16.38 = -16.35 + -0.03)
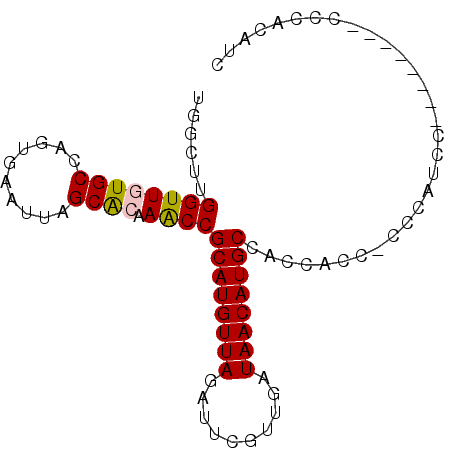
| Location | 14,622,689 – 14,622,779 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -19.50 |
| Energy contribution | -20.50 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
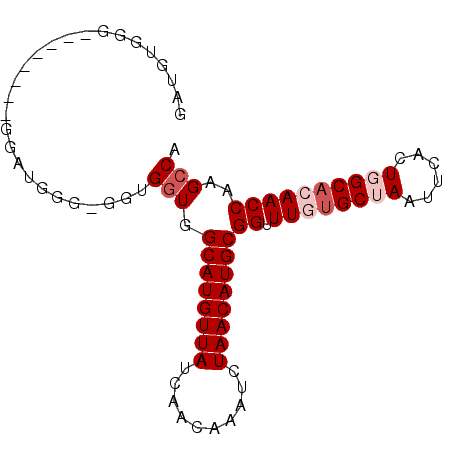
>3R_DroMel_CAF1 14622689 90 - 27905053 GAUGUGGGCGGACCAAGGAUGGGCGUUGGUGGCAUGUUAUCAACGAAUCUAACAUGCGGUUUGUGCUAAUUCACUGGCACAACCAAGCCA ......(((..(((((.(.....).))))).((((((((..........))))))))((((.((((((......))))))))))..))). ( -32.30) >DroPse_CAF1 35650 72 - 1 GAUGUUGG------------------GGGUGGCAUGUUAUCAACAAAUCUAACAUGCGGCUUGUGCUAAUUCACUCGCAGAACCAAGCCA ....(((.------------------(((((((((((((..........))))))))(((....)))....))))).))).......... ( -20.60) >DroEre_CAF1 36275 80 - 1 GCUGU----------AGGAUGGGAGGUGCUGGCAUGUUAUCAACGAAUCUAACAUGCGGUUUGUGCUAAUUCACUGGCACAACCGAGCCA .....----------.........(((....((((((((..........))))))))((((.((((((......))))))))))..))). ( -24.80) >DroYak_CAF1 38091 90 - 1 UAUGUGGGCGGCACAAGGAUGGGAGGCGGUGGCAUGUUAUCAACGAAUCUAACAUGCGGUUUGCGCUAAUUCACUGGCACAACCAAGCCA ..((((.....)))).........(((....((((((((..........))))))))((((.(.((((......)))).)))))..))). ( -26.00) >DroAna_CAF1 35122 80 - 1 --AGUGGG--------GGAGUCGCGGUGGUGGCAUGUUAUCAACAAAUCUAACAUGCGGUUUGUGCUAAUUCACUGGCACAACCAAGCCA --.((((.--------....))))(((....((((((((..........))))))))((((.((((((......))))))))))..))). ( -26.90) >DroPer_CAF1 38304 71 - 1 GAUGUUGG-------------------GGUGGCAUGUUAUCAACAAAUCUAACAUGCGGCUUGUGCUAAUUCACUCGCAGAACCAAGCCA ........-------------------....((((((((..........))))))))(((((((((..........)))....)))))). ( -18.70) >consensus GAUGUGGG________GGAUGGG_GGUGGUGGCAUGUUAUCAACAAAUCUAACAUGCGGUUUGUGCUAAUUCACUGGCACAACCAAGCCA ...........................(((.((((((((..........))))))))((.((((((((......))))))))))..))). (-19.50 = -20.50 + 1.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:34 2006