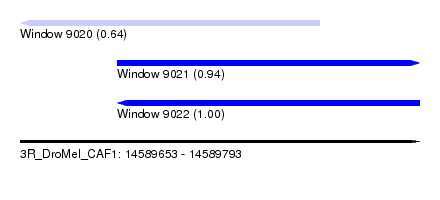
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,589,653 – 14,589,793 |
| Length | 140 |
| Max. P | 0.998006 |

| Location | 14,589,653 – 14,589,758 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -17.69 |
| Consensus MFE | -8.95 |
| Energy contribution | -9.75 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14589653 105 - 27905053 UUUCUUUAUCAAUUUCCACAACAAGCUC------AAGCUC---AGAGCUUAGUCCUGGGAAAAGCAAGAAGAGCAAGAAAAUCUAGAAAAAACCAAAGAACACGAAUUAGAUGA ((((((......((((((..((((((((------......---.)))))).))..))))))..((.......))))))))((((((....................)))))).. ( -18.95) >DroSec_CAF1 4386 94 - 1 UUUCUUUAUCAAUUUCCACAACAAGCUCAAGCUCAAGCUC---AGAGCUUAGUCCUGGGAAAAGCAAGC-----------------AAAAAACCAAAGAACACGCAUUAGAUGA .....(((((..((((((..((((((((.(((....))).---.)))))).))..))))))......((-----------------.................))....))))) ( -20.33) >DroSim_CAF1 4316 93 - 1 UUUCUUUAUCAAUUUCCACAACAAGCUCAAGCUCAAGCUC---AGAGCUUAGUCCUGGUAAAAGCAAG------------------AAAAAACCAAAGAACACGCAUUAGAUGA ((((((.........(((..((((((((.(((....))).---.)))))).))..))).......)))------------------)))......................... ( -16.99) >DroEre_CAF1 4276 106 - 1 UUUCUUUAUCAAUUUCCACA-CAAGCUC------AGGCUCAGAGGAGCUUAGUGCUGGGAAAAGCAGCAAAAACAAGAAAAACACG-AAAAACCCAAGAACACCAAAUAGAUGA .....(((((..((((((((-(((((((------..........)))))).))).)))))).........................-..........(.....).....))))) ( -17.10) >DroYak_CAF1 5226 94 - 1 UUUCUUUAUCAAUUUCCACA-CAAGCUC------AAGCUC---GGAGCUUAGUGCUGAGAAAAGAAGCAAAAACAAGAA----------AAACCAAAGAACACAAAAUAGAUGA .((((((.....((((((((-(((((((------......---.)))))).))).)).))))..............(..----------....))))))).............. ( -15.10) >consensus UUUCUUUAUCAAUUUCCACAACAAGCUC______AAGCUC___AGAGCUUAGUCCUGGGAAAAGCAAGAA_A_CAAGAA_______AAAAAACCAAAGAACACGAAUUAGAUGA .....(((((..((((((..((((((((................)))))).))..))))))....................................(....)......))))) ( -8.95 = -9.75 + 0.80)

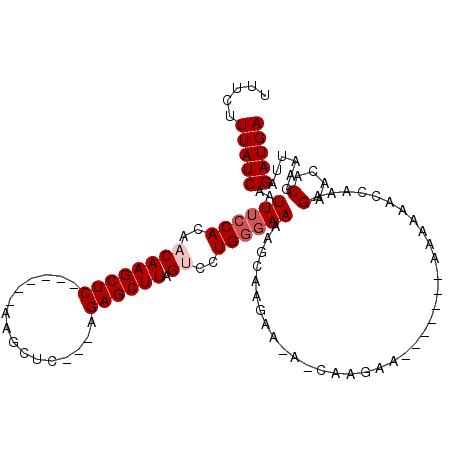
| Location | 14,589,687 – 14,589,793 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.29 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -19.00 |
| Energy contribution | -18.82 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14589687 106 + 27905053 UUUCUUGCUCUUCUUGCUUUUCCCAGGACUAAGCUCU---GAGCUU------GAGCUUGUUGUGGAAAUUGAUAAAGAAAUAUUAUUGUAGGAUAAAAUAUCCCACAAGCAGAGA (((((.((..(((((.(..((.(((.(((.((((((.---......------))))))))).))).))..)...)))))......((((.(((((...))))).))))))))))) ( -30.80) >DroSec_CAF1 4414 99 + 1 -----------GCUUGCUUUUCCCAGGACUAAGCUCU---GAGCUUGAGCUUGAGCUUGUUGUGGAAAUUGAUAAAGAAAUAUUAUUGUAGGAUACAAUAUCCCACAAGGAGA-- -----------.(((.(..((.(((.(((.((((((.---((((....))))))))))))).))).))..)...)))........((((.(((((...))))).)))).....-- ( -28.40) >DroEre_CAF1 4309 106 + 1 UUUCUUGUUUUUGCUGCUUUUCCCAGCACUAAGCUCCUCUGAGCCU------GAGCUUG-UGUGGAAAUUGAUAAAGAAAUAUUAUUGUAGGAUACAAUAUCCCAGAAGCAGA-- ....(((((((((.(((.(((((..((((.((((((..........------)))))))-)))))))).(((((......)))))..)))(((((...)))))))))))))).-- ( -25.30) >DroYak_CAF1 5251 102 + 1 -UUCUUGUUUUUGCUUCUUUUCUCAGCACUAAGCUCC---GAGCUU------GAGCUUG-UGUGGAAAUUGAUAAAGAAAUAUUAUUGUAGGAUAGAAUAUCCCACAAGAAGA-- -(((.(((((((...((.(((((..((((.((((((.---......------)))))))-))))))))..))...)))))))...((((.(((((...))))).)))))))..-- ( -25.60) >consensus _UUCUUGUUUUUCCUGCUUUUCCCAGCACUAAGCUCC___GAGCUU______GAGCUUG_UGUGGAAAUUGAUAAAGAAAUAUUAUUGUAGGAUACAAUAUCCCACAAGCAGA__ .............((((.(((((((....(((((((................))))))).)).))))).................((((.(((((...))))).))))))))... (-19.00 = -18.82 + -0.19)

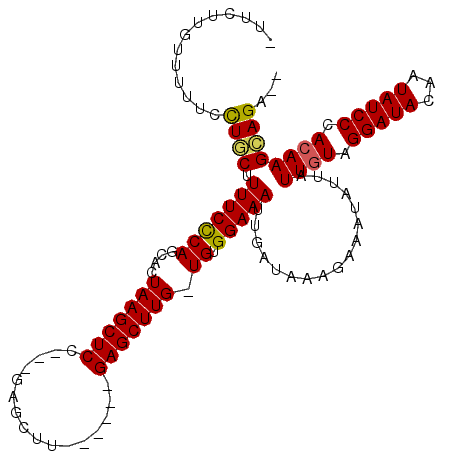
| Location | 14,589,687 – 14,589,793 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.29 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -19.14 |
| Energy contribution | -20.64 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
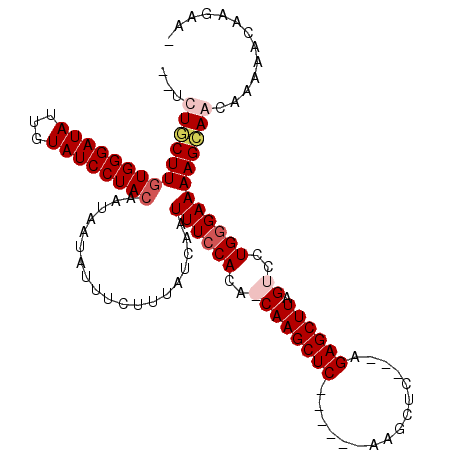
>3R_DroMel_CAF1 14589687 106 - 27905053 UCUCUGCUUGUGGGAUAUUUUAUCCUACAAUAAUAUUUCUUUAUCAAUUUCCACAACAAGCUC------AAGCUC---AGAGCUUAGUCCUGGGAAAAGCAAGAAGAGCAAGAAA (((.(((((((((((((...))))))))........(((((......((((((..((((((((------......---.)))))).))..))))))....))))))))))))).. ( -31.60) >DroSec_CAF1 4414 99 - 1 --UCUCCUUGUGGGAUAUUGUAUCCUACAAUAAUAUUUCUUUAUCAAUUUCCACAACAAGCUCAAGCUCAAGCUC---AGAGCUUAGUCCUGGGAAAAGCAAGC----------- --....((((((..(((((((........)))))))..)........((((((..((((((((.(((....))).---.)))))).))..))))))..))))).----------- ( -28.40) >DroEre_CAF1 4309 106 - 1 --UCUGCUUCUGGGAUAUUGUAUCCUACAAUAAUAUUUCUUUAUCAAUUUCCACA-CAAGCUC------AGGCUCAGAGGAGCUUAGUGCUGGGAAAAGCAGCAAAAACAAGAAA --.((((((..(..(((((((........)))))))..)........((((((((-(((((((------..........)))))).))).))))))))))))............. ( -29.30) >DroYak_CAF1 5251 102 - 1 --UCUUCUUGUGGGAUAUUCUAUCCUACAAUAAUAUUUCUUUAUCAAUUUCCACA-CAAGCUC------AAGCUC---GGAGCUUAGUGCUGAGAAAAGAAGCAAAAACAAGAA- --.((((((((((((((...)))))))))..................((((((((-(((((((------......---.)))))).))).)).)))))))))............- ( -26.90) >consensus __UCUGCUUGUGGGAUAUUGUAUCCUACAAUAAUAUUUCUUUAUCAAUUUCCACA_CAAGCUC______AAGCUC___AGAGCUUAGUCCUGGGAAAAGCAACAAAAACAAGAA_ ...((((((((((((((...))))))))...................((((((..((((((((................)))))).))..))))))))))))............. (-19.14 = -20.64 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:26 2006