| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,052,639 – 2,052,768 |
| Length | 129 |
| Max. P | 0.767380 |

| Location | 2,052,639 – 2,052,757 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.43 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -10.42 |
| Energy contribution | -11.38 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
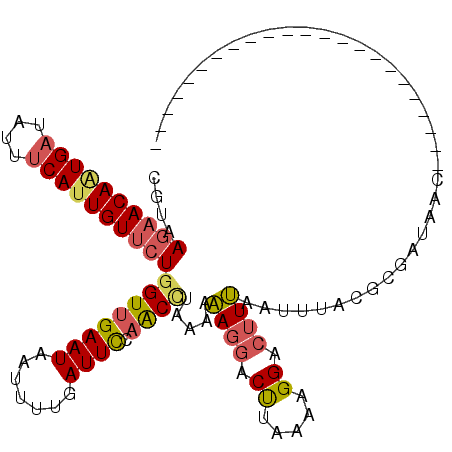
>3R_DroMel_CAF1 2052639 118 + 27905053 CUUUUACAUAAGGC--CGAUUGAAAUUGAAAUUUGGUAACCGUAAGAACAGUGACAUGUCAUUGUUCUUGUUGAAUAAUUUUGAUUCCAGCCAGAAAAAGGACUUAAAAGGACUUUAUCU ...........(((--....((.(((..(((((...((((...(((((((((((....)))))))))))))))...)))))..))).)))))(((.((((..((....))..)))).))) ( -31.90) >DroSec_CAF1 196104 118 + 1 CUUUUACUUAACGC--CCUCUUAAAUUCAAAUUUGGUAACCGUAACAACAAUGAUAUUUCAUUGUUCUGGUUGAAUAAUUUUGAUUCCAACCCAAAAAAGGACUUAAAAGGACUUUAAUU .......((((.(.--(((.((((.(((...(((((((((((....((((((((....)))))))).)))))).......(((....))).)))))...))).)))).))).).)))).. ( -21.40) >DroSim_CAF1 201093 118 + 1 CUUUUACAUAACGC--CCUCUUAAAUUCAAAUUUGGUAACCGUAAGAACAAUGAUAUUUCAUUGUUCUGGUUGAAUAAUUUUGAUUCCAACCUAAAAAAGGACUUAAAAGGACUUUAAUU ..............--(((.((((..(((((.(((.(((((...((((((((((....)))))))))))))))..))).)))))......(((.....)))..)))).)))......... ( -25.40) >DroEre_CAF1 195991 116 + 1 CGUUUGCAUAAGAC--UGACUUAAAUACAGAUUUGUUGG-CGUAAGAACAAUGAUAUACCACUGUGCUGGUCGAAUAGUUUCGAUUUAAGCUUAAAACAGCACCUAAAUGGAUAUGUA-G ....((((((.(.(--((........((((..(..(((.-(....)..)))..).......))))(((.((((((....))))))...)))......)))).((.....)).))))))-. ( -21.50) >DroYak_CAF1 199435 95 + 1 CUUUUACAUAAGACGUCGAUUUAAAUAUAGAAUUGUUAGCCAUAAGAACAAUGAAAUAUCAUUGUGCUGCUUGCCUAGUUUCGAUAUGAGUCAAA------------------------- .................((((((.((...(((...((((.((..((.(((((((....))))))).))...)).)))).))).)).))))))...------------------------- ( -17.10) >consensus CUUUUACAUAAGGC__CGACUUAAAUUCAAAUUUGGUAACCGUAAGAACAAUGAUAUAUCAUUGUUCUGGUUGAAUAAUUUUGAUUCCAGCCAAAAAAAGGACUUAAAAGGACUUUAA_U .......................(((..(((((...(((((...((((((((((....)))))))))))))))...)))))..))).................................. (-10.42 = -11.38 + 0.96)


| Location | 2,052,639 – 2,052,757 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.43 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -7.88 |
| Energy contribution | -8.40 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2052639 118 - 27905053 AGAUAAAGUCCUUUUAAGUCCUUUUUCUGGCUGGAAUCAAAAUUAUUCAACAAGAACAAUGACAUGUCACUGUUCUUACGGUUACCAAAUUUCAAUUUCAAUCG--GCCUUAUGUAAAAG (((.((((..((....))..)))).)))((((((.................(((((((.(((....))).)))))))..((...))...............)))--)))........... ( -23.20) >DroSec_CAF1 196104 118 - 1 AAUUAAAGUCCUUUUAAGUCCUUUUUUGGGUUGGAAUCAAAAUUAUUCAACCAGAACAAUGAAAUAUCAUUGUUGUUACGGUUACCAAAUUUGAAUUUAAGAGG--GCGUUAAGUAAAAG ..((((.((((((((((((.(...(((((((((((..........)))))))..((((((((....))))))))...........))))...).))))))))))--)).))))....... ( -31.30) >DroSim_CAF1 201093 118 - 1 AAUUAAAGUCCUUUUAAGUCCUUUUUUAGGUUGGAAUCAAAAUUAUUCAACCAGAACAAUGAAAUAUCAUUGUUCUUACGGUUACCAAAUUUGAAUUUAAGAGG--GCGUUAUGUAAAAG .......((((((((((((.(.......(((((((..........)))))))((((((((((....))))))))))................).))))))))))--))............ ( -31.60) >DroEre_CAF1 195991 116 - 1 C-UACAUAUCCAUUUAGGUGCUGUUUUAAGCUUAAAUCGAAACUAUUCGACCAGCACAGUGGUAUAUCAUUGUUCUUACG-CCAACAAAUCUGUAUUUAAGUCA--GUCUUAUGCAAACG .-........(((....((((((.((((....))))(((((....))))).)))))).)))(((((..((((..((((..-...(((....)))...)))).))--))..)))))..... ( -19.20) >DroYak_CAF1 199435 95 - 1 -------------------------UUUGACUCAUAUCGAAACUAGGCAAGCAGCACAAUGAUAUUUCAUUGUUCUUAUGGCUAACAAUUCUAUAUUUAAAUCGACGUCUUAUGUAAAAG -------------------------...(((((((((.(((..(((.((...((.(((((((....))))))).))..)).)))....))).)))).......)).)))........... ( -13.91) >consensus A_UUAAAGUCCUUUUAAGUCCUUUUUUAGGCUGGAAUCAAAAUUAUUCAACCAGAACAAUGAAAUAUCAUUGUUCUUACGGUUACCAAAUUUGAAUUUAAGUCG__GCCUUAUGUAAAAG ....................................................((((((((((....))))))))))............................................ ( -7.88 = -8.40 + 0.52)

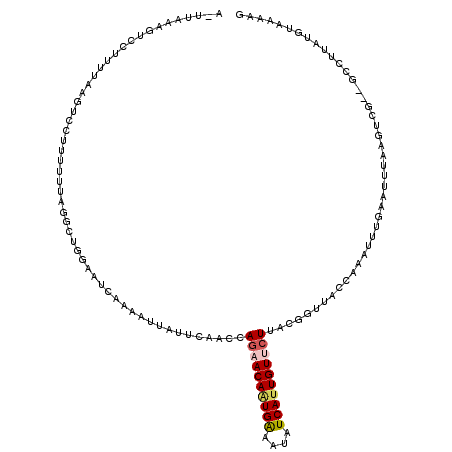
| Location | 2,052,677 – 2,052,768 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.62 |
| Mean single sequence MFE | -20.62 |
| Consensus MFE | -10.88 |
| Energy contribution | -11.62 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2052677 91 + 27905053 CGUAAGAACAGUGACAUGUCAUUGUUCUUGUUGAAUAAUUUUGAUUCCAGCCAGAAAAAGGACUUAAAAGGACUUUAUCUUACGCGAUAAC----------------------- ..((((((((((((....))))))))))))...................((.(((.((((..((....))..)))).)))...))......----------------------- ( -22.30) >DroSec_CAF1 196142 114 + 1 CGUAACAACAAUGAUAUUUCAUUGUUCUGGUUGAAUAAUUUUGAUUCCAACCCAAAAAAGGACUUAAAAGGACUUUAAUUUACGCGAUAACAACUUUUUGGACAGUGCUUAAAC .((((.((((((((....))))))))..((((((((.......))).)))))....((((..((....))..))))...))))(((....(((....))).....)))...... ( -19.60) >DroSim_CAF1 201131 114 + 1 CGUAAGAACAAUGAUAUUUCAUUGUUCUGGUUGAAUAAUUUUGAUUCCAACCUAAAAAAGGACUUAAAAGGACUUUAAUUUACGCGAUAACAACUUUUUGGACAGUGCCUAAAC ....((((((((((....))))))))))((((((((.......))).)))))....((((..((....))..))))..((((.(((....(((....))).....))).)))). ( -24.30) >DroEre_CAF1 196028 82 + 1 CGUAAGAACAAUGAUAUACCACUGUGCUGGUCGAAUAGUUUCGAUUUAAGCUUAAAACAGCACCUAAAUGGAUAUGUA-G-AAU------------------------------ .............((((((((..((((((((((((....))))))(((....)))..)))))).....))).))))).-.-...------------------------------ ( -16.30) >consensus CGUAAGAACAAUGAUAUUUCAUUGUUCUGGUUGAAUAAUUUUGAUUCCAACCUAAAAAAGGACUUAAAAGGACUUUAAUUUACGCGAUAAC_______________________ ....((((((((((....))))))))))((((((((.......)))).)))).....((((.((.....)).))))...................................... (-10.88 = -11.62 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:54 2006