
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
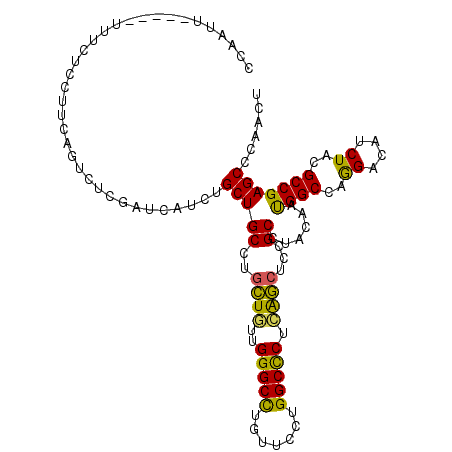
| Location | 14,521,048 – 14,521,153 |
| Length | 105 |
| Max. P | 0.809525 |

| Location | 14,521,048 – 14,521,153 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.45 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -23.27 |
| Energy contribution | -22.42 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14521048 105 + 27905053 CCCAUU-----UUUUUCCUUUAGUUUCGAUCAUCUGCUGCCUGCUGUUGGGCCUGUUCCUGGCCCUCAGCUCCGCCUACAAUGGCCAAGACAUCUAUGCCGAGCCCAAUU ......-----........................(((((..((((..(((((.......))))).))))...))......((((..((....))..)))))))...... ( -25.30) >DroSec_CAF1 17852 105 + 1 CCCAUU-----UUUCUCCUUCAGUUUCGAUCAUCUGCUGCCUGCUGUUGGGCCUGUUCCUGGCCCUCAGCUCCGCCUACAAUGGCCAGGACAUCUAUGCCGAGCCCAAUU ......-----.............((((..(((..(.(((((((((..(((((.......))))).))))...(((......))).))).)).).))).))))....... ( -27.80) >DroSim_CAF1 17805 105 + 1 CCCAUU-----UUUCUCCUUCAGUUUCGAUCAUCUGCUGCCUGCUGUUGGGCCUGUUCCUGGCCCUCAGCUCCGCCUACAAUGGCCAGGACAUCUAUGCCGAGCCCAAUU ......-----.............((((..(((..(.(((((((((..(((((.......))))).))))...(((......))).))).)).).))).))))....... ( -27.80) >DroEre_CAF1 17432 110 + 1 CAAUUUUCAAUUUUCUCCUGCAGUCUCCAUCAUUUGCUGCCUGCUCUUGGGCCUCUUCCUGGCCCUCAGCUCCGCCUACAAUGGCCAGGACAUCUACGCCGAGCCCAACU ...................((((.(..((.....))..).))))..((((((....((((((((..................))))))))..((......)))))))).. ( -27.97) >DroYak_CAF1 24008 105 + 1 CAAAUU-----UUCCUUCUGCAGUCUCCCUCAUUUGCUGCCUACUGUUGGGCUUGUUCCUGGCCCUCAGCUCCGCCUACAAUGGCCAGGACAUCUACGCCGAGCCCAACU ......-----........(((((...........))))).....((((((((((.((((((((..................)))))))).........)))))))))). ( -32.87) >DroAna_CAF1 16791 107 + 1 UUAAUU---UUUUGCCCCUUUAGGCACUCUCCUCUGCUGCCUGGUUCUGGGCCUGAUCCUGGCUCUGGCCAGCGCCUACAACGGCCAGGACAUCUACGCCGAGCCCAACU ......---............((((((........).)))))(((((..(((..((((((((((..(((....)))......))))))))..))...))))))))..... ( -33.00) >consensus CCAAUU_____UUUCUCCUUCAGUCUCGAUCAUCUGCUGCCUGCUGUUGGGCCUGUUCCUGGCCCUCAGCUCCGCCUACAAUGGCCAGGACAUCUACGCCGAGCCCAACU ...................................(((((..((((..(((((.......))))).))))...))......((((..((....))..)))))))...... (-23.27 = -22.42 + -0.86)


| Location | 14,521,048 – 14,521,153 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.45 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -24.86 |
| Energy contribution | -24.78 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14521048 105 - 27905053 AAUUGGGCUCGGCAUAGAUGUCUUGGCCAUUGUAGGCGGAGCUGAGGGCCAGGAACAGGCCCAACAGCAGGCAGCAGAUGAUCGAAACUAAAGGAAAAA-----AAUGGG ..((((..(((((((...((((((.(((......))).))((((.(((((.......)))))..)))).))))....))).))))..))))........-----...... ( -35.20) >DroSec_CAF1 17852 105 - 1 AAUUGGGCUCGGCAUAGAUGUCCUGGCCAUUGUAGGCGGAGCUGAGGGCCAGGAACAGGCCCAACAGCAGGCAGCAGAUGAUCGAAACUGAAGGAGAAA-----AAUGGG ..(..(..(((((((...((.(((.(((......)))...((((.(((((.......)))))..)))))))))....))).))))..)..)........-----...... ( -33.80) >DroSim_CAF1 17805 105 - 1 AAUUGGGCUCGGCAUAGAUGUCCUGGCCAUUGUAGGCGGAGCUGAGGGCCAGGAACAGGCCCAACAGCAGGCAGCAGAUGAUCGAAACUGAAGGAGAAA-----AAUGGG ..(..(..(((((((...((.(((.(((......)))...((((.(((((.......)))))..)))))))))....))).))))..)..)........-----...... ( -33.80) >DroEre_CAF1 17432 110 - 1 AGUUGGGCUCGGCGUAGAUGUCCUGGCCAUUGUAGGCGGAGCUGAGGGCCAGGAAGAGGCCCAAGAGCAGGCAGCAAAUGAUGGAGACUGCAGGAGAAAAUUGAAAAUUG .((((.(((((((.(.....((((((((....(((......)))..))))))))..).)))...))))...))))...((..(....)..)).................. ( -33.60) >DroYak_CAF1 24008 105 - 1 AGUUGGGCUCGGCGUAGAUGUCCUGGCCAUUGUAGGCGGAGCUGAGGGCCAGGAACAAGCCCAACAGUAGGCAGCAAAUGAGGGAGACUGCAGAAGGAA-----AAUUUG .(((((((((......))((((((((((....(((......)))..))))))).))).))))))).....(((((..........).))))........-----...... ( -35.80) >DroAna_CAF1 16791 107 - 1 AGUUGGGCUCGGCGUAGAUGUCCUGGCCGUUGUAGGCGCUGGCCAGAGCCAGGAUCAGGCCCAGAACCAGGCAGCAGAGGAGAGUGCCUAAAGGGGCAAAA---AAUUAA .....(((((((((....)))).((((((.((....)).)))))))))))........((((......(((((.(......)..)))))....))))....---...... ( -35.60) >consensus AAUUGGGCUCGGCAUAGAUGUCCUGGCCAUUGUAGGCGGAGCUGAGGGCCAGGAACAGGCCCAACAGCAGGCAGCAGAUGAUCGAAACUGAAGGAGAAA_____AAUGGG ..((((((((......))..((((((((......(((...)))...))))))))....)))))).............................................. (-24.86 = -24.78 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:08 2006