
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,515,084 – 14,515,232 |
| Length | 148 |
| Max. P | 0.992209 |

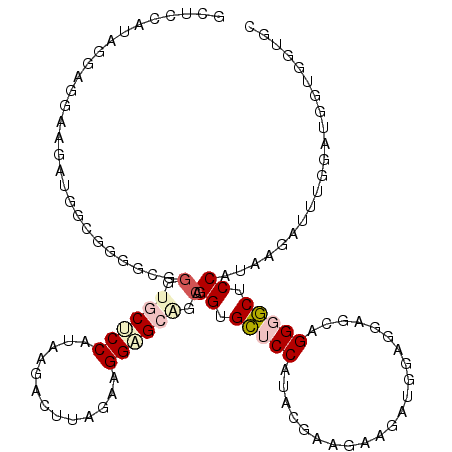
| Location | 14,515,084 – 14,515,200 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.20 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -13.06 |
| Energy contribution | -14.04 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14515084 116 + 27905053 GCUCCGUAGGAGGAAGAUGGCGGGGCGGGUGCUCCAUAAGACUUGGAAGGAGCAGCAGGAGCUCCAUACGAAGAAGAUGGAGGAGCAGGGGCUCCAUAAGAUUUGGAUGGUGGUGC (((((((............))))))).(.((((((.............)))))).)....((.((((.((((....((((((.........))))))....))))....)))).)) ( -38.52) >DroPse_CAF1 11699 116 + 1 GCUCCAUAAGAUGAUGAUGGCUGCGGGGGUGCACCAUACGAUUUGGAUGGUGGUGCAGGAGCUCCAUAAGAUGAUGAUGGCUGCUGGGGGGCACCAUACGAUUUCGAUGGUGGUGC (((((...((..........))...)))))(((((((.(((..(.(((((((.(.(((.((((.(((......)))..)))).)))...).)))))).).)..)))...))))))) ( -38.60) >DroSec_CAF1 12035 98 + 1 ------------------GGCGGGGCGGGUGCUCCAUAAGACUUAGAAGGAGCAGCAGGUGCUCCAUACGAAGAAGAUGGAGGAGCAGGGGCUCCGUAAGAUUUGGAUGGUGGUGC ------------------.(((((((.(.((((((.............)))))).).(.(.((((((.(......))))))).).)....)))))))................... ( -33.92) >DroEre_CAF1 11393 116 + 1 GCUCCGUAGGAGGAAGAUGGUGGUGCAGGUGCUCCGUAGGAGGAAGACGGUGGGGCAGGUGCUCCAUAAGACUUGGAAGGAGCAGCAGGCGCUCCAUACGAAGAAGAUGGAGGAGC .((((((.........((((.(((((.(.((((((....(((.......(((((((....)))))))....)))....)))))).)..))))))))).........)))))).... ( -44.97) >DroYak_CAF1 11211 97 + 1 GCUCCAUAGGAGGAAGAUGGUGGGGCCGGUGCUCCAUAAGACUUGGAAGGAGCAGCGGGGGUUCCAUACGAAGAAGAGGGAGGAGCUGGGACUCCAU------------------- .((((...)))).......(((((((((.((((((.............)))))).))).((((((...(........)...))))))....))))))------------------- ( -33.62) >DroAna_CAF1 11104 116 + 1 GCACCAUAGGAGGGUGAUGGCGGGGCAGGUGCUCCAUAAGACUUAGAUGGCGCUGGAGGUGCUCCAUAUGAUGGAGAAGGAGCGGCAGGUGCCCCAUAUGAUUGGGAUGGUGGGGC .((((((.....(((.(((..(((((((.((((((..............((((.....))))(((((...)))))...)))))).)...)))))).))).)))...)))))).... ( -42.10) >consensus GCUCCAUAGGAGGAAGAUGGCGGGGCGGGUGCUCCAUAAGACUUAGAAGGAGCAGCAGGUGCUCCAUACGAAGAAGAUGGAGGAGCAGGGGCUCCAUAAGAUUUGGAUGGUGGUGC ...........................(.((((((.............)))))).).((.(((((......................))))).))..................... (-13.06 = -14.04 + 0.97)


| Location | 14,515,084 – 14,515,200 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.20 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -9.58 |
| Energy contribution | -10.64 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14515084 116 - 27905053 GCACCACCAUCCAAAUCUUAUGGAGCCCCUGCUCCUCCAUCUUCUUCGUAUGGAGCUCCUGCUGCUCCUUCCAAGUCUUAUGGAGCACCCGCCCCGCCAUCUUCCUCCUACGGAGC .....................(((((....)))))........(((((((.((((.......((((((.............))))))...((...)).......))))))))))). ( -28.52) >DroPse_CAF1 11699 116 - 1 GCACCACCAUCGAAAUCGUAUGGUGCCCCCCAGCAGCCAUCAUCAUCUUAUGGAGCUCCUGCACCACCAUCCAAAUCGUAUGGUGCACCCCCGCAGCCAUCAUCAUCUUAUGGAGC ((((((....(((....(.((((((.....(((.(((..((((......)))).))).)))...)))))))....)))..))))))......((..((((.........)))).)) ( -26.40) >DroSec_CAF1 12035 98 - 1 GCACCACCAUCCAAAUCUUACGGAGCCCCUGCUCCUCCAUCUUCUUCGUAUGGAGCACCUGCUGCUCCUUCUAAGUCUUAUGGAGCACCCGCCCCGCC------------------ ....................(((.((...((((((........(((.(...((((((.....))))))..).)))......))))))...)).)))..------------------ ( -26.74) >DroEre_CAF1 11393 116 - 1 GCUCCUCCAUCUUCUUCGUAUGGAGCGCCUGCUGCUCCUUCCAAGUCUUAUGGAGCACCUGCCCCACCGUCUUCCUCCUACGGAGCACCUGCACCACCAUCUUCCUCCUACGGAGC ((((((((((...(((.(...((((((.....))))))..).)))....)))))(((..(((....((((.........)))).)))..)))...................))))) ( -31.80) >DroYak_CAF1 11211 97 - 1 -------------------AUGGAGUCCCAGCUCCUCCCUCUUCUUCGUAUGGAACCCCCGCUGCUCCUUCCAAGUCUUAUGGAGCACCGGCCCCACCAUCUUCCUCCUAUGGAGC -------------------..(((((....)))))............(.((((.....(((.((((((.............)))))).))).....)))))((((......)))). ( -26.32) >DroAna_CAF1 11104 116 - 1 GCCCCACCAUCCCAAUCAUAUGGGGCACCUGCCGCUCCUUCUCCAUCAUAUGGAGCACCUCCAGCGCCAUCUAAGUCUUAUGGAGCACCUGCCCCGCCAUCACCCUCCUAUGGUGC ((((((..............)))))).......(((((.............))))).......(((((((...((..(.((((.((....))....)))).)..))...))))))) ( -33.06) >consensus GCACCACCAUCCAAAUCGUAUGGAGCCCCUGCUCCUCCAUCUUCUUCGUAUGGAGCACCUGCUGCUCCUUCCAAGUCUUAUGGAGCACCCGCCCCGCCAUCUUCCUCCUAUGGAGC .....................(((((....)))))..............((((..........(((((.............)))))..........))))................ ( -9.58 = -10.64 + 1.06)



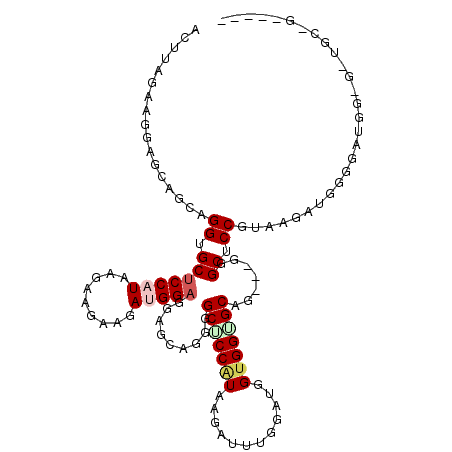
| Location | 14,515,124 – 14,515,232 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.75 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -13.34 |
| Energy contribution | -14.18 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14515124 108 + 27905053 ACUUGGAAGGAGCAGCAGGAGCUCCAUACGAAGAAGAUGGAGGAGCAGGGGCUCCAUAAGAUUUGGAUGGUGGUGCUG---GGGCUCCGUAAGAUGGGGAUGGCGGUGCAG----- .((((...(((((..(((.(.(.((((.((((....((((((.........))))))....)))).)))).).).)))---..))))).))))..................----- ( -34.50) >DroPse_CAF1 11739 113 + 1 AUUUGGAUGGUGGUGCAGGAGCUCCAUAAGAUGAUGAUGGCUGCUGGGGGGCACCAUACGAUUUCGAUGGUGGUGCAG---GUGCUCCAUAAGAAGAGGAAGGUGCUGCUGGUCUU ....((((((..(..(((.((((.(((......)))..)))).))..((((((((.(((.(((.....))).)))..)---)))))))..............)..)..)).)))). ( -34.70) >DroSec_CAF1 12057 95 + 1 ACUUAGAAGGAGCAGCAGGUGCUCCAUACGAAGAAGAUGGAGGAGCAGGGGCUCCGUAAGAUUUGGAUGGUGGUGCUG---GGGCUCCGUAAGAUGGG------------------ .((((...(((((....((..((((((.((((....((((((.........))))))....)))).)))).))..)).---..))))).)))).....------------------ ( -33.50) >DroEre_CAF1 11433 108 + 1 AGGAAGACGGUGGGGCAGGUGCUCCAUAAGACUUGGAAGGAGCAGCAGGCGCUCCAUACGAAGAAGAUGGAGGAGCAG---GGGCUCCAUAAGAUUUUGAUGGCGGUGCGG----- ......((.(((((((....))))).(((((((((...(((((..(..((.((((((.(......)))))))..))..---).))))).)))).)))))...)).))....----- ( -35.50) >DroAna_CAF1 11144 103 + 1 ACUUAGAUGGCGCUGGAGGUGCUCCAUAUGAUGGAGAAGGAGCGGCAGGUGCCCCAUAUGAUUGGGAUGGUGGGGCGGGAGGAGCUCCGUAUGACUUCGAUGG------------- ...............(((((.((((((...))))))..(((((......((((((((............))))))))......))))).....))))).....------------- ( -34.00) >consensus ACUUAGAAGGAGCAGCAGGUGCUCCAUAAGAAGAAGAUGGAGGAGCAGGGGCUCCAUAAGAUUUGGAUGGUGGUGCAG___GGGCUCCGUAAGAUGGGGAUGG_G_UGC_G_____ .................((.(((((((.........))))).........(((((((............))))))).......)).))............................ (-13.34 = -14.18 + 0.84)


| Location | 14,515,124 – 14,515,232 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.75 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -10.68 |
| Energy contribution | -11.64 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14515124 108 - 27905053 -----CUGCACCGCCAUCCCCAUCUUACGGAGCCC---CAGCACCACCAUCCAAAUCUUAUGGAGCCCCUGCUCCUCCAUCUUCUUCGUAUGGAGCUCCUGCUGCUCCUUCCAAGU -----.......................(((((..---.((((..................(((((....)))))((((((......).))))).....)))))))))........ ( -23.60) >DroPse_CAF1 11739 113 - 1 AAGACCAGCAGCACCUUCCUCUUCUUAUGGAGCAC---CUGCACCACCAUCGAAAUCGUAUGGUGCCCCCCAGCAGCCAUCAUCAUCUUAUGGAGCUCCUGCACCACCAUCCAAAU ((((...((((....((((.........))))...---))))..((((((((....)).))))))....................)))).(((.((....)).))).......... ( -23.40) >DroSec_CAF1 12057 95 - 1 ------------------CCCAUCUUACGGAGCCC---CAGCACCACCAUCCAAAUCUUACGGAGCCCCUGCUCCUCCAUCUUCUUCGUAUGGAGCACCUGCUGCUCCUUCUAAGU ------------------.....((((.(((((..---.((((..................(((((....)))))((((((......).))))).....)))))))))...)))). ( -24.00) >DroEre_CAF1 11433 108 - 1 -----CCGCACCGCCAUCAAAAUCUUAUGGAGCCC---CUGCUCCUCCAUCUUCUUCGUAUGGAGCGCCUGCUGCUCCUUCCAAGUCUUAUGGAGCACCUGCCCCACCGUCUUCCU -----..(((.(((..............(((((..---..)))))((((((......).))))))))..)))((((((.............))))))................... ( -26.02) >DroAna_CAF1 11144 103 - 1 -------------CCAUCGAAGUCAUACGGAGCUCCUCCCGCCCCACCAUCCCAAUCAUAUGGGGCACCUGCCGCUCCUUCUCCAUCAUAUGGAGCACCUCCAGCGCCAUCUAAGU -------------...............((.(((......((((((..............)))))).......(((((.............)))))......))).))........ ( -25.26) >consensus _____C_GCA_C_CCAUCCACAUCUUACGGAGCCC___CAGCACCACCAUCCAAAUCGUAUGGAGCCCCUGCUCCUCCAUCUUCAUCGUAUGGAGCACCUGCAGCACCAUCUAAGU ............................((((........(((((................)))))........)))).............((.((.......)).))........ (-10.68 = -11.64 + 0.96)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:05 2006