
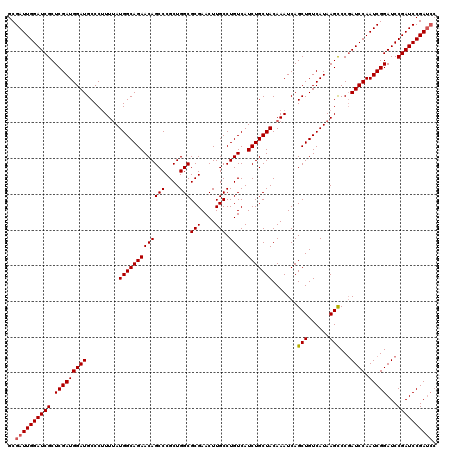
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,469,474 – 14,469,673 |
| Length | 199 |
| Max. P | 0.745406 |

| Location | 14,469,474 – 14,469,594 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -37.75 |
| Energy contribution | -38.20 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14469474 120 + 27905053 GCGAUUGGAUCGCUCGAUGGAUGCCCUUUUAUGGCAGAACAGCCCGCUGGCGCGAACUUGCCUGUCAUCUGCUACAAAUCAGCUGUCAUAAGUCCGAUCCAAUCGGAUCCGAUCCGAUCC ..((((((((((..(....(((((...(((.(((((((((((((....)))(((....))).)))..))))))).)))...)).)))....)..))))))))))(((((......))))) ( -41.50) >DroSim_CAF1 27341 120 + 1 GCGAUUGGAUCGCUCGAUGGAUGCCCUUUUAUGGCAGAACAGCCCGCUGGCGCGAACUUGCCUGUCAUCUGCUACAAAUCAGCUGUCAUAAGCCCGAUCCAAUCGGAUCCGAUCCGAUCC ..((((((((((((..((((..((...(((.(((((((((((((....)))(((....))).)))..))))))).)))...))..)))).)))..(((((....))))).))))))))). ( -41.80) >DroYak_CAF1 30167 117 + 1 GCGAUUGGAUCGCUCGAUGGAUGCCCGUUAAUGGCAGAACAGCCCGCUGGCGCGAACUUGCCUGUCAUCUGCUACAAAUCAGCUGUCAUAAGCCCCAUCCAAUCGGAUCCGAUCCGA--- (.(((((((((...(((((((((...((..(((((((....(((....)))(((....))))))))))..)).........(((......)))..))))).))))))))))))))..--- ( -40.20) >consensus GCGAUUGGAUCGCUCGAUGGAUGCCCUUUUAUGGCAGAACAGCCCGCUGGCGCGAACUUGCCUGUCAUCUGCUACAAAUCAGCUGUCAUAAGCCCGAUCCAAUCGGAUCCGAUCCGAUCC ..((((((((((.(((((((((.........(((((((((((((....)))(((....))).)))..))))))).......(((......)))...)))).)))))...)))))))))). (-37.75 = -38.20 + 0.45)


| Location | 14,469,474 – 14,469,594 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -40.27 |
| Energy contribution | -41.93 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14469474 120 - 27905053 GGAUCGGAUCGGAUCCGAUUGGAUCGGACUUAUGACAGCUGAUUUGUAGCAGAUGACAGGCAAGUUCGCGCCAGCGGGCUGUUCUGCCAUAAAAGGGCAUCCAUCGAGCGAUCCAAUCGC (((((......)))))((((((((((..((((((...(((..(((((.(((((......(((.((((((....)))))))))))))).)))))..)))...))).))))))))))))).. ( -47.10) >DroSim_CAF1 27341 120 - 1 GGAUCGGAUCGGAUCCGAUUGGAUCGGGCUUAUGACAGCUGAUUUGUAGCAGAUGACAGGCAAGUUCGCGCCAGCGGGCUGUUCUGCCAUAAAAGGGCAUCCAUCGAGCGAUCCAAUCGC (((((......)))))(((((((((..(((((((...(((..(((((.(((((......(((.((((((....)))))))))))))).)))))..)))...))).))))))))))))).. ( -48.10) >DroYak_CAF1 30167 117 - 1 ---UCGGAUCGGAUCCGAUUGGAUGGGGCUUAUGACAGCUGAUUUGUAGCAGAUGACAGGCAAGUUCGCGCCAGCGGGCUGUUCUGCCAUUAACGGGCAUCCAUCGAGCGAUCCAAUCGC ---...(((.(((((.(.((.((((((((((......((((.....)))).((((.((((..(((((((....)))))))..)))).))))...)))).)))))).)))))))).))).. ( -42.80) >consensus GGAUCGGAUCGGAUCCGAUUGGAUCGGGCUUAUGACAGCUGAUUUGUAGCAGAUGACAGGCAAGUUCGCGCCAGCGGGCUGUUCUGCCAUAAAAGGGCAUCCAUCGAGCGAUCCAAUCGC (((((......)))))((((((((((..((((((...(((..(((((.(((((......(((.((((((....)))))))))))))).)))))..)))...))).))))))))))))).. (-40.27 = -41.93 + 1.67)


| Location | 14,469,554 – 14,469,673 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -36.97 |
| Consensus MFE | -29.34 |
| Energy contribution | -30.01 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14469554 119 - 27905053 UCAAAGAUUGUGUGGCUGUUAGUUGGGUUUUUGAAAGUGAUUGUAACAAUCGCCGAAAUAAUGCUGCCC-GACUCACAUUGGAUCGGAUCGGAUCCGAUUGGAUCGGACUUAUGACAGCU .............((((((((((((((((((((...(((((((...)))))))))))).......))))-))).....((.((((.(((((....))))).)))).))....)))))))) ( -39.71) >DroSim_CAF1 27421 120 - 1 UCAAAGAUUGUGUGGCUGUUAGUUGGGUUUUUGAAAGUGAUUGUAACAAUCGCCGAAAUAAUGCUGCCCGGACCCACAUUGGAUCGGAUCGGAUCCGAUUGGAUCGGGCUUAUGACAGCU .............((((((((((.....(((((...(((((((...))))))))))))....)).((((((..(((.((((((((......))))))))))).))))))...)))))))) ( -40.70) >DroYak_CAF1 30247 110 - 1 UCAAAGAUUAUGUUGCUCUUAGUUGGGGCUUUGAAAGUGAUUGUAACAAUCGCCGAAAUAAUGCUUCCCCAAC----------UCGGAUCGGAUCCGAUUGGAUGGGGCUUAUGACAGCU .......(((((..(((((..(((((((.((((...(((((((...))))))))))).........)))))))----------((.(((((....))))).)).))))).)))))..... ( -30.50) >consensus UCAAAGAUUGUGUGGCUGUUAGUUGGGUUUUUGAAAGUGAUUGUAACAAUCGCCGAAAUAAUGCUGCCC_GAC_CACAUUGGAUCGGAUCGGAUCCGAUUGGAUCGGGCUUAUGACAGCU ..............(((((((((((((((((((...(((((((...)))))))))))).......)))).)))........((((.(((((....))))).)))).......))))))). (-29.34 = -30.01 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:46 2006