| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,048,437 – 2,048,542 |
| Length | 105 |
| Max. P | 0.959418 |

| Location | 2,048,437 – 2,048,542 |
|---|---|
| Length | 105 |
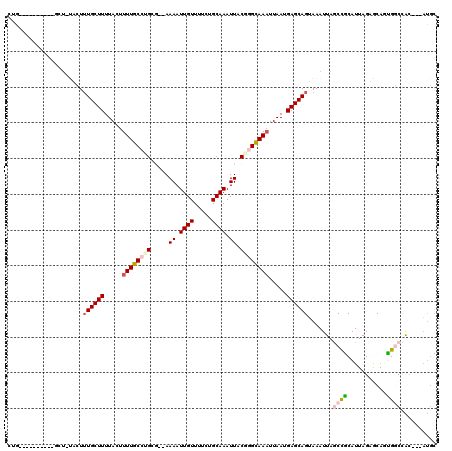
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.27 |
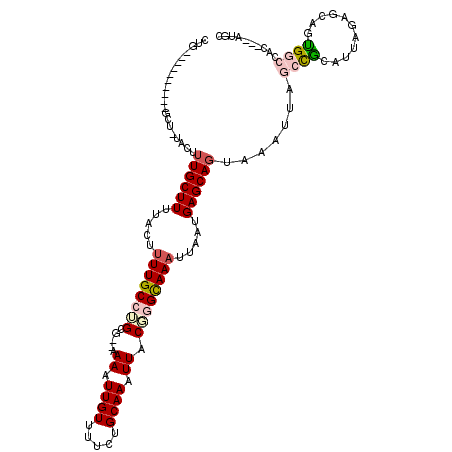
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -14.22 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2048437 105 - 27905053 CGA----------GCUGCACUUUGCUUUUACUUUUGCCUGCG--AAAAUUGUUUUUAGCAAAUUGCGGGCAAAUUAAUGAGCAGUAAAUUAGCCGCAUUAGUACGGCGGCCAC---AUGC ...----------...(((..((((((.....((((((((((--(...((((.....)))).))))))))))).....)))))).......(((((.........)))))...---.))) ( -33.60) >DroVir_CAF1 240604 104 - 1 CUG----------GCU-UUGUUUGCUUUUACUUUUGCCUGC---AAAAUUGUUUUCUGCAAAUUACUCGCAAAUUAAUGAGCAGAAAAUUAACCACAUUAGAGCACUGCCCGCUC--CCU ..(----------((.-.(((((........(((((....)---))))..(((((((((..((((.........))))..)))))))))...........)))))..))).....--... ( -20.80) >DroEre_CAF1 191492 99 - 1 ---------------UGCACUUUGCUUUUACUUUUGCCUGCG--AAAAUUGUU-CCAGCAAAUUGCGGGCAAAUUAAUGAGCAGUAAAUUAGCUGCAUUAGUGCAGCGGCCAC---AUGC ---------------.((...((((((.....((((((((((--(...((((.-...)))).))))))))))).....)))))).......((((((....)))))).))...---.... ( -35.20) >DroWil_CAF1 192114 116 - 1 GUGUGUGUGUUUUUGU-UUUUAUGCUUUUACUUUUGCCAGAGUAAAAAUUGUUUUCUGCAAAUUACAGGCAAAUUAAUGAGCAGUAAAUUAGCCGCAUUAGAGCAGUGGCCAC---AUGC (((((((.((..((((-(((.((((((((((((......)))))))).((((.....))))......(((.((((.((.....)).)))).))))))).)))))))..)))))---)))) ( -32.10) >DroMoj_CAF1 196511 106 - 1 CUG----------GCU-UUGUUUGCUUUUACUUUUGCCAGC---AAAAUUGUUUUGUGCAAAUUACUCGCAAUUUAAUGAGCAGAAAAUUAACCACAUUAGAGCACUGCCUGGCUAAUGU .((----------(((-..((.((((((...((((((((..---.(((((((...(((.....)))..)))))))..)).)))))).............))))))..))..))))).... ( -21.49) >DroAna_CAF1 168409 115 - 1 CAGUAUGUGCUUUGCCUUACUUUGCUUUUACUUUUGCCUGCG--AAAAUUGUUUUGUGCAAAUUGCGGGUAAAUUAAUGAGCAGUAAAUUAGCCGCAUUAGUACAAUGGCCAC---AUGC ..(((((((.......(((((..((((.....((((((((((--(...((((.....)))).))))))))))).....)))))))))....((((...........)))))))---)))) ( -32.60) >consensus CUG__________GCU_UACUUUGCUUUUACUUUUGCCUGCG__AAAAUUGUUUUCUGCAAAUUACGGGCAAAUUAAUGAGCAGUAAAUUAGCCGCAUUAGAGCAGUGGCCAC___AUGC .....................((((((.....((((((((.....((.((((.....)))).)).)))))))).....)))))).......((((...........)))).......... (-14.22 = -15.00 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:50 2006