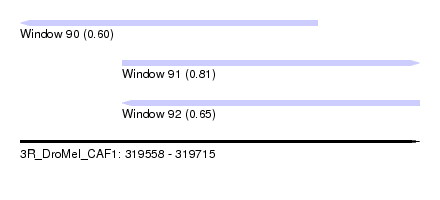
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 319,558 – 319,715 |
| Length | 157 |
| Max. P | 0.808342 |

| Location | 319,558 – 319,675 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -44.35 |
| Consensus MFE | -28.59 |
| Energy contribution | -29.45 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
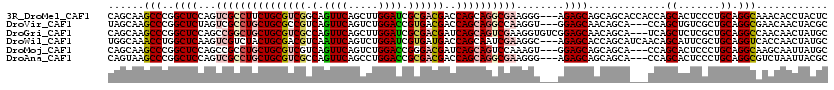
>3R_DroMel_CAF1 319558 117 - 27905053 CAGCAAGCCCGGCUCCAGUCGCCUUCUGCGUCGGCAGUUCAGCUUGGAUCGCGACGACCAGCAGGCGAAGGG---AGAGCAGCAGCACCACCAGCACUCCCUGCAGGCAAACACCUACUC ......(((...((((..((((((.(((.((((((.((((.....)))).))..))))))).))))))..))---)).((((..((.......)).....)))).)))............ ( -42.20) >DroVir_CAF1 4349 114 - 1 UAGCAAGCCCGGCUCUAGUCGCCUGCUGCGCCGUCAGUUCAGUCUGGACCGUGACGACCAGCAGGCCAAGGU---GGAGCAACAGCA---CCAGCUGUCGCUGCAGGCGAACAACUACGC ..(((.((...((((((.(.((((((((.(.(((((((((.....))))..))))).)))))))))...).)---))))).(((((.---...))))).)))))..(((........))) ( -46.80) >DroGri_CAF1 5053 117 - 1 CAGCAAGCCCGGCUCCAGCCGGCUGCUGCGUCGCCAGUUCAGCUUGGAUCGCGACGAUCAGCAGUCGAAGGUGUCGGAGCAACAGCA---UCAGCUCUCGCUGCAGGCCAACAACUAUGC ..(((.(((((((.((...(((((((((((((((..((((.....)))).))))))..)))))))))..)).))))).))....((.---.((((....))))...)).........))) ( -49.70) >DroWil_CAF1 9032 117 - 1 UGGCAAACCUGGCUCAAGUCGUCUACUGCGACGUCAAUUCAGUCUGGAUCGUGAUGACCAGCAAUCGAAGGC---AGAGCACCAGCAUCAACAGCAUUCGCUGCAGGUCACCAACUAUGC (((...(((((((((..(((((.....)))))(((..(((.((((((.((.....))))))..)).))))))---.))))...........((((....)))))))))..)))....... ( -33.20) >DroMoj_CAF1 491 114 - 1 CAGCAAGCCCGGCUCCAGCCGCCUGCUGCGUCGUCAGUUCAGUCUGGACCGGGACGAUCAGCAGUCCAAAGU---GGAGCAGCAGCA---CCAGCACUCCCUGCAGGCAAGCAAUUAUGC ..((..(((.(((....)))((((((((.((((((.((((.....))))...)))))))))))).....((.---((((..((....---...)).)))))))).)))..))........ ( -41.10) >DroAna_CAF1 7502 114 - 1 CAGUAAGCCCGGCUCCAGUCGCCUGCUGCGUCGCCAGUUCAGCCUGGACCGCGACGACCAGCAGGCGAAGGG---AGAGCAGCAGCA---CCAGCACUCCCUGCAGGCGUCUAAUUACGC ..((((....(((.((..((((((((((((((((..((((.....)))).))))))..))))))))))((((---((.((.......---...)).))))))...)).)))...)))).. ( -53.10) >consensus CAGCAAGCCCGGCUCCAGUCGCCUGCUGCGUCGUCAGUUCAGCCUGGACCGCGACGACCAGCAGGCGAAGGU___AGAGCAGCAGCA___CCAGCACUCCCUGCAGGCAAACAACUACGC ......(((..((((...(((((((((((((((.(.((((.....)))).))))))..))))))))))........)))).............((.......)).)))............ (-28.59 = -29.45 + 0.86)
| Location | 319,598 – 319,715 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -42.73 |
| Consensus MFE | -30.67 |
| Energy contribution | -30.81 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
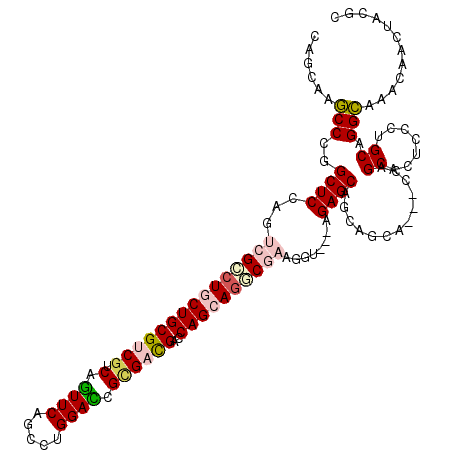
>3R_DroMel_CAF1 319598 117 + 27905053 GCUCU---CCCUUCGCCUGCUGGUCGUCGCGAUCCAAGCUGAACUGCCGACGCAGAAGGCGACUGGAGCCGGGCUUGCUGUCGAACCGAGAGGCUCCUCCCCCACACCCACUUCCGCUCA ((.((---((..((((((((((((((...))).)).)))....((((....)))).))))))..))))..(((..((..(..((.((....)).))..)...))..)))......))... ( -37.90) >DroVir_CAF1 4386 100 + 1 GCUCC---ACCUUGGCCUGCUGGUCGUCACGGUCCAGACUGAACUGACGGCGCAGCAGGCGACUAGAGCCGGGCUUGCUAUCAAACUUUGAGGC---------------GCUGCCACU-- ((((.---....(.(((((((((((((((((((....))))...))))))).)))))))).)...))))..(((.((((.(((.....))))))---------------)..)))...-- ( -43.60) >DroPse_CAF1 471 100 + 1 GUUCU---CCCUUCGCCUGCUGGUCGUCGCGAUCGAGACUGAACUGGCGACGCAGCAGGCGACUGGAGCCGGGUUUGCUAUCGAAUUUCGCCG-U--------------ACUGCCGCC-- ...((---((..(((((((((((((((((.(.(((....))).)))))))).))))))))))..))))...(((.(((...((.....))..)-)--------------)..)))...-- ( -42.40) >DroGri_CAF1 5090 103 + 1 GCUCCGACACCUUCGACUGCUGAUCGUCGCGAUCCAAGCUGAACUGGCGACGCAGCAGCCGGCUGGAGCCGGGCUUGCUGUCAAACUUGGAGGU---------------GCUGCCGGC-- ...(((.(((((((((((((((..((((((.(((......))..).))))))))))))(((((....)))))..............))))))))---------------)....))).-- ( -41.30) >DroAna_CAF1 7539 114 + 1 GCUCU---CCCUUCGCCUGCUGGUCGUCGCGGUCCAGGCUGAACUGGCGACGCAGCAGGCGACUGGAGCCGGGCUUACUGUCGUACCGAUUGACUCCUC---CAAUUCCGAUGCCGCUUC ((.((---((..((((((((((((((((.((((....))))....)))))).))))))))))..))))...(((.....((((....(((((.......---))))).)))))))))... ( -48.80) >DroPer_CAF1 471 100 + 1 GUUCU---CCCUUCGCCUGCUGGUCGUCGCGAUCGAGACUGAACUGGCGACGCAGCAGGCGACUGGAGCCGGGUUUGCUAUCGAAUUUCGCCG-U--------------ACUGCCGCC-- ...((---((..(((((((((((((((((.(.(((....))).)))))))).))))))))))..))))...(((.(((...((.....))..)-)--------------)..)))...-- ( -42.40) >consensus GCUCU___CCCUUCGCCUGCUGGUCGUCGCGAUCCAGACUGAACUGGCGACGCAGCAGGCGACUGGAGCCGGGCUUGCUAUCGAACUUAGAGGCU______________ACUGCCGCC__ (((((.......(((((((((((((((((.(.((......)).)))))))).))))))))))..)))))..(((......................................)))..... (-30.67 = -30.81 + 0.14)


| Location | 319,598 – 319,715 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -44.47 |
| Consensus MFE | -28.12 |
| Energy contribution | -28.40 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 319598 117 - 27905053 UGAGCGGAAGUGGGUGUGGGGGAGGAGCCUCUCGGUUCGACAGCAAGCCCGGCUCCAGUCGCCUUCUGCGUCGGCAGUUCAGCUUGGAUCGCGACGACCAGCAGGCGAAGGG---AGAGC ...((....))((((.((..(...(((((....)))))..)..)).))))..((((..((((((.(((.((((((.((((.....)))).))..))))))).))))))..))---))... ( -49.60) >DroVir_CAF1 4386 100 - 1 --AGUGGCAGC---------------GCCUCAAAGUUUGAUAGCAAGCCCGGCUCUAGUCGCCUGCUGCGCCGUCAGUUCAGUCUGGACCGUGACGACCAGCAGGCCAAGGU---GGAGC --.(.(((.((---------------...(((.....)))..))..)))).((((((.(.((((((((.(.(((((((((.....))))..))))).)))))))))...).)---))))) ( -39.00) >DroPse_CAF1 471 100 - 1 --GGCGGCAGU--------------A-CGGCGAAAUUCGAUAGCAAACCCGGCUCCAGUCGCCUGCUGCGUCGCCAGUUCAGUCUCGAUCGCGACGACCAGCAGGCGAAGGG---AGAAC --((..((.((--------------.-(((......))))).))....))..((((..((((((((((((((((....((......))..))))))..))))))))))..))---))... ( -42.20) >DroGri_CAF1 5090 103 - 1 --GCCGGCAGC---------------ACCUCCAAGUUUGACAGCAAGCCCGGCUCCAGCCGGCUGCUGCGUCGCCAGUUCAGCUUGGAUCGCGACGAUCAGCAGUCGAAGGUGUCGGAGC --.(((...((---------------((((.(..(((((....)))))..(((....)))((((((((((((((..((((.....)))).))))))..))))))))).)))))))))... ( -44.20) >DroAna_CAF1 7539 114 - 1 GAAGCGGCAUCGGAAUUG---GAGGAGUCAAUCGGUACGACAGUAAGCCCGGCUCCAGUCGCCUGCUGCGUCGCCAGUUCAGCCUGGACCGCGACGACCAGCAGGCGAAGGG---AGAGC ...(((((((((..((((---(.....)))))))))((....))..)))...((((..((((((((((((((((..((((.....)))).))))))..))))))))))..))---)).)) ( -49.60) >DroPer_CAF1 471 100 - 1 --GGCGGCAGU--------------A-CGGCGAAAUUCGAUAGCAAACCCGGCUCCAGUCGCCUGCUGCGUCGCCAGUUCAGUCUCGAUCGCGACGACCAGCAGGCGAAGGG---AGAAC --((..((.((--------------.-(((......))))).))....))..((((..((((((((((((((((....((......))..))))))..))))))))))..))---))... ( -42.20) >consensus __AGCGGCAGC______________AGCCUCGAAGUUCGACAGCAAGCCCGGCUCCAGUCGCCUGCUGCGUCGCCAGUUCAGCCUGGAUCGCGACGACCAGCAGGCGAAGGG___AGAGC ...............................................((((((....)))((((((((((((((..((((.....)))).))))))..))))))))...)))........ (-28.12 = -28.40 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:35 2006