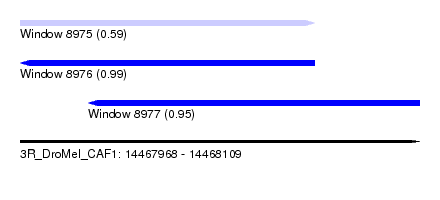
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,467,968 – 14,468,109 |
| Length | 141 |
| Max. P | 0.989459 |

| Location | 14,467,968 – 14,468,072 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -25.44 |
| Energy contribution | -24.33 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14467968 104 + 27905053 ----------------AAUGUAUAAACACCUAAUUUUGGUAAGUGUUUUGGCCAAGAGCAUCUUGCCUGCAAGCCGUUCUGCAGUUGACCUUGCGUUGCAGGUCUUUGGCUGUCACAGAA ----------------...........(((.......)))..(((...(((((((((((..((((....))))..)))))......(((((.((...)))))))..)))))).))).... ( -28.50) >DroSim_CAF1 25862 120 + 1 UUCGAUAUGUAAUGAAAAUGUAUAAACACCCAAUUUUGGUGAGUGUUUUGGCCAAGAGCAUCUUGCCUGCAAGCCGUUCUGCAGUUGACCUUGCGUUGCAGGUCUUUGGCUGUCACAGAA .((.......................((((.......)))).(((...(((((((((((..((((....))))..)))))......(((((.((...)))))))..)))))).))).)). ( -30.70) >DroYak_CAF1 28222 109 + 1 -----------AUGAAAAUGUAUCAACACCUUAUUUUUCUGAGUGUUUUAGCCAAGAGCAUCUUGCCUGCAAACCGUUCUGCAGCUGACUUUGUUUUGCAAGUCCUUGGCUGCCACAGAA -----------.......(((...(((((.(((......)))))))).((((((((.((.....))(((((........)))))..(((((.(.....))))))))))))))..)))... ( -24.30) >consensus ___________AUGAAAAUGUAUAAACACCUAAUUUUGGUGAGUGUUUUGGCCAAGAGCAUCUUGCCUGCAAGCCGUUCUGCAGUUGACCUUGCGUUGCAGGUCUUUGGCUGUCACAGAA ..........................................(((...((((((((.((.....))(((((........)))))..(((((.((...))))))))))))))).))).... (-25.44 = -24.33 + -1.10)

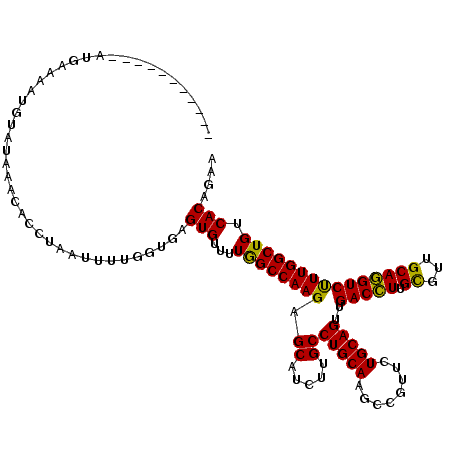
| Location | 14,467,968 – 14,468,072 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -28.40 |
| Energy contribution | -28.73 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14467968 104 - 27905053 UUCUGUGACAGCCAAAGACCUGCAACGCAAGGUCAACUGCAGAACGGCUUGCAGGCAAGAUGCUCUUGGCCAAAACACUUACCAAAAUUAGGUGUUUAUACAUU---------------- ...((((...(((((.(((((((...)).)))))..((((((......))))))((.....))..)))))..(((((((((.......))))))))).))))..---------------- ( -30.60) >DroSim_CAF1 25862 120 - 1 UUCUGUGACAGCCAAAGACCUGCAACGCAAGGUCAACUGCAGAACGGCUUGCAGGCAAGAUGCUCUUGGCCAAAACACUCACCAAAAUUGGGUGUUUAUACAUUUUCAUUACAUAUCGAA ...(((((..(((((.(((((((...)).)))))..((((((......))))))((.....))..)))))..(((((((((.......)))))))))...........)))))....... ( -33.30) >DroYak_CAF1 28222 109 - 1 UUCUGUGGCAGCCAAGGACUUGCAAAACAAAGUCAGCUGCAGAACGGUUUGCAGGCAAGAUGCUCUUGGCUAAAACACUCAGAAAAAUAAGGUGUUGAUACAUUUUCAU----------- (((((.((((((....(((((........))))).)))))......((((..((.((((.....)))).)).)))).).)))))....(((((((....)))))))...----------- ( -31.30) >consensus UUCUGUGACAGCCAAAGACCUGCAACGCAAGGUCAACUGCAGAACGGCUUGCAGGCAAGAUGCUCUUGGCCAAAACACUCACCAAAAUUAGGUGUUUAUACAUUUUCAU___________ ...((((...(((((.(((((........)))))..((((((......))))))((.....))..)))))..(((((((((.......))))))))).)))).................. (-28.40 = -28.73 + 0.34)

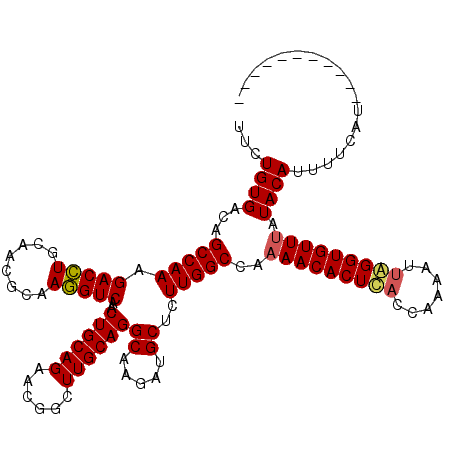
| Location | 14,467,992 – 14,468,109 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -34.33 |
| Energy contribution | -33.67 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14467992 117 - 27905053 CUGGCUUGCCAGAAGAAGCC---UCUUCGACCAAGUCACAUUCUGUGACAGCCAAAGACCUGCAACGCAAGGUCAACUGCAGAACGGCUUGCAGGCAAGAUGCUCUUGGCCAAAACACUU ((((....))))((((.((.---((((.......(((((.....))))).(((...(((((((...)).)))))...(((((......)))))))))))).))))))............. ( -37.20) >DroSim_CAF1 25902 117 - 1 ---GUUUGCCAGAAGAAGCCUCCUCUUCGACCAAGUCACAUUCUGUGACAGCCAAAGACCUGCAACGCAAGGUCAACUGCAGAACGGCUUGCAGGCAAGAUGCUCUUGGCCAAAACACUC ---(((((((((((((.......)))))((.((.(((((.....))))).(((...(((((((...)).)))))...(((((......))))))))....)).)).))))..)))).... ( -36.80) >DroYak_CAF1 28251 114 - 1 ---GUGUGCCAGAAGAAGCC---UCUUCGACCAAGUCACAUUCUGUGGCAGCCAAGGACUUGCAAAACAAAGUCAGCUGCAGAACGGUUUGCAGGCAAGAUGCUCUUGGCUAAAACACUC ---(((((((((((((....---)))))(((...)))........))))((((((((((((........)))))..(((((((....)))))))((.....)).)))))))...)))).. ( -40.00) >consensus ___GUUUGCCAGAAGAAGCC___UCUUCGACCAAGUCACAUUCUGUGACAGCCAAAGACCUGCAACGCAAGGUCAACUGCAGAACGGCUUGCAGGCAAGAUGCUCUUGGCCAAAACACUC .......(((((((((.......)))))((.((.(((((.....))))).(((...(((((........)))))...(((((......))))))))....)).)).)))).......... (-34.33 = -33.67 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:42 2006