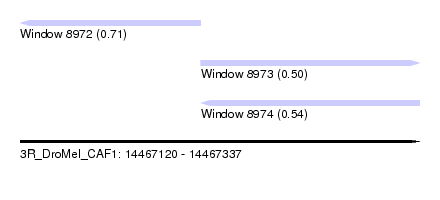
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,467,120 – 14,467,337 |
| Length | 217 |
| Max. P | 0.711227 |

| Location | 14,467,120 – 14,467,218 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.19 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -17.69 |
| Energy contribution | -18.80 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
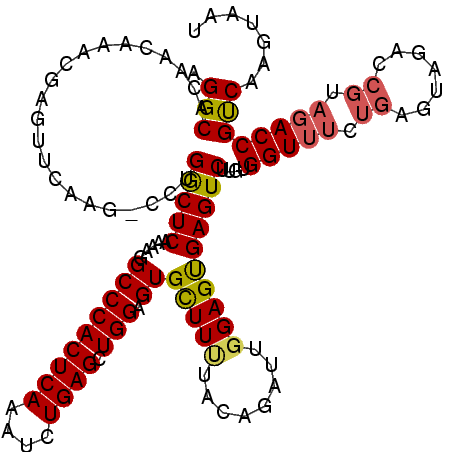
>3R_DroMel_CAF1 14467120 98 - 27905053 ------CGCAUUCAACAACAAUUGCAGCCAGUUGAGGUUGAAGCGUCUGGUUGAACGGUUGUUGUACUUGUCUGGCGAUCAGAGAAGCCCAAGAAU----------------AACAAAAG ------(((.((((((..((((((....))))))..))))))))).(((......)))(((((((.((((...(((..........))))))).))----------------)))))... ( -28.80) >DroSim_CAF1 25048 79 - 1 ------CGUAUUCAACAACAAUUGCAGUCAGUUGAGGUUGAAGCGUCUGGUUGAAAGGUUGUUGU-------------------AAGCCCAAGAAU----------------AACUAAAG ------(((.((((((..((((((....))))))..)))))))))..((((((...((((.....-------------------.))))......)----------------)))))... ( -18.20) >DroYak_CAF1 26833 120 - 1 CGUGUUCGUAUUCAACAACAAUUGCAGUCAGUUGAGGUUGAAGCGUCUGGUUGAAUGGUUGUUGUACUUGUCUGGCGAUCAGAAAAGCCCAAGAAUUACUACAGACCGUAAUAAUAAAAG ..((((((..((((((..((((((....))))))..))))))..(((((....(.(((((.(((..(((.((((.....)))).)))..))).))))).).))))))).))))....... ( -29.30) >consensus ______CGUAUUCAACAACAAUUGCAGUCAGUUGAGGUUGAAGCGUCUGGUUGAAAGGUUGUUGUACUUGUCUGGCGAUCAGA_AAGCCCAAGAAU________________AACAAAAG ......(((.((((((..((((((....))))))..)))))))))............(((.(((..(((.((((.....)))).)))..))).)))........................ (-17.69 = -18.80 + 1.11)


| Location | 14,467,218 – 14,467,337 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.57 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -21.55 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14467218 119 + 27905053 AUUACUUGACGGUCUACGGUCUACCCAGAAACCAAAAGACUCACUCCAAUCUGGAAAAGCACUCCAGCUCAGAUUUGAGUGGGCCUUUUGAGUCAGA-CUUGAACUCGUUUGUUGUGCCU ..(((..((((((.((.(((((.....((...((((((.(((((((.((((((....(((......))))))))).))))))).))))))..)))))-)))).)).))))....)))... ( -33.50) >DroSim_CAF1 25127 120 + 1 AUUACUUGGCGGUCUACGGUCUAUUCAGAAACCACAAGACUCACUCGAAUCUGAAAAAGCACUCCAGCUCAGAUUUGAGUGGGCCUUUUGAGCCAAGUUUCGAACUCGUUUGUUGUGCCU ...(((((((((.(....).))..........((.(((.(((((((((((((((....((......))))))))))))))))).))).)).)))))))....(((......)))...... ( -39.30) >DroYak_CAF1 26953 112 + 1 AAUACUUGACGGU-------CUACUCAGAAACCACAAGACUCGCUCCACUCUGUAGAAACACUCCAUCUCAGAUUUGAGUGGGCUUUUUGAGCCAGG-CCUGAACUAGUUUGUCGUGCCU .......((((((-------((.(((((((......((.(((((((...((((.(((.........)))))))...))))))))))))))))..)))-)).(((....))))))...... ( -28.90) >consensus AUUACUUGACGGUCUACGGUCUACUCAGAAACCACAAGACUCACUCCAAUCUGAAAAAGCACUCCAGCUCAGAUUUGAGUGGGCCUUUUGAGCCAGG_CUUGAACUCGUUUGUUGUGCCU ....(((((((((....((.....))....)))..(((.(((((((.((((((................)))))).))))))).)))....))))))....................... (-21.55 = -22.22 + 0.68)

| Location | 14,467,218 – 14,467,337 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -23.99 |
| Energy contribution | -24.67 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14467218 119 - 27905053 AGGCACAACAAACGAGUUCAAG-UCUGACUCAAAAGGCCCACUCAAAUCUGAGCUGGAGUGCUUUUCCAGAUUGGAGUGAGUCUUUUGGUUUCUGGGUAGACCGUAGACCGUCAAGUAAU .(((.........((((.((..-..))))))(((((((.(((((.(((((((((......)))....)))))).))))).)))))))(((((.(((.....))).))))))))....... ( -39.40) >DroSim_CAF1 25127 120 - 1 AGGCACAACAAACGAGUUCGAAACUUGGCUCAAAAGGCCCACUCAAAUCUGAGCUGGAGUGCUUUUUCAGAUUCGAGUGAGUCUUGUGGUUUCUGAAUAGACCGUAGACCGCCAAGUAAU ............((....))..(((((((....(((((.(((((.((((((((..((....))..)))))))).))))).)))))..(((((.((.......)).))))))))))))... ( -39.00) >DroYak_CAF1 26953 112 - 1 AGGCACGACAAACUAGUUCAGG-CCUGGCUCAAAAAGCCCACUCAAAUCUGAGAUGGAGUGUUUCUACAGAGUGGAGCGAGUCUUGUGGUUUCUGAGUAG-------ACCGUCAAGUAUU ......(((...(((.((((((-((.(((((.....(((((((((....)))).))).))(((((........))))))))))....)))..))))))))-------...)))....... ( -31.10) >consensus AGGCACAACAAACGAGUUCAAG_CCUGGCUCAAAAGGCCCACUCAAAUCUGAGCUGGAGUGCUUUUACAGAUUGGAGUGAGUCUUGUGGUUUCUGAGUAGACCGUAGACCGUCAAGUAAU .(((......................(((((.....(((((((((....)))).))).))(((((........))))))))))....(((((.((.......)).))))))))....... (-23.99 = -24.67 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:39 2006