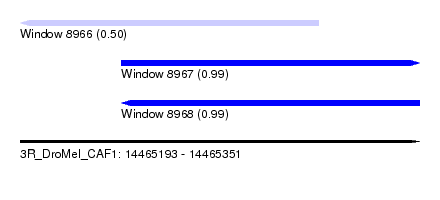
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,465,193 – 14,465,351 |
| Length | 158 |
| Max. P | 0.991300 |

| Location | 14,465,193 – 14,465,311 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -38.24 |
| Energy contribution | -38.13 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14465193 118 - 27905053 AGUUUUGGCCACUUGGCGCAUGCGUAACUUUUUUGCUGGC--GCUGGGAAGAGCCAGCCAGUUGCAAAAUGCGCAGUCCGACACGACACCAUGCGAAAUAUUCGAGUGUUUUAUUGGCCA .....((((((....((((((..((((((.....((((((--.((....)).)))))).))))))...))))))..........(((((....(((.....))).)))))....)))))) ( -43.00) >DroSim_CAF1 23079 120 - 1 AGUUUGGGCCACUUGGCGCAUGCGUAACUUUUUUGCUGGCGCGCUGGGAAGAGUCAGCCAGUUGCAAAAUGCGCAGUCCGACACGACACCAUGCGAAAUAUUCGAAUGUUUUAUUGGCCA ......(((((....((((((..((((((.....((((((...((....)).)))))).))))))...)))))).(((......)))..(((.(((.....))).)))......))))). ( -38.70) >DroYak_CAF1 24871 116 - 1 AGAUUUGGCCACUUGGCGCAUGCGUAACUU-CUUGCUGGC--GCUGGGAAGAGCCA-CAAGUUGCACAAUGCGCAGUCCGACACGACACCAUGCGAAGUAUUCGAGUGUUUUAUUGGCCG ......(((((....(((((((.((((((.-..((.((((--.((....)).))))-)))))))).).))))))..........(((((....(((.....))).)))))....))))). ( -39.00) >consensus AGUUUUGGCCACUUGGCGCAUGCGUAACUUUUUUGCUGGC__GCUGGGAAGAGCCAGCCAGUUGCAAAAUGCGCAGUCCGACACGACACCAUGCGAAAUAUUCGAGUGUUUUAUUGGCCA ......(((((....((((((..((((((.....((((((...((....)).)))))).))))))...)))))).(((......)))..(((.(((.....))).)))......))))). (-38.24 = -38.13 + -0.11)


| Location | 14,465,233 – 14,465,351 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -44.77 |
| Consensus MFE | -32.73 |
| Energy contribution | -33.40 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14465233 118 + 27905053 CGGACUGCGCAUUUUGCAACUGGCUGGCUCUUCCCAGC--GCCAGCAAAAAAGUUACGCAUGCGCCAAGUGGCCAAAACUAUGGCCACCCGAUGGAAAGCCAGCCAGCAGACCGACCGAC (((.((((((.....))...(((((((((.((((..((--((..((...........))..))))...(((((((......))))))).....))))))))))))))))).)))...... ( -53.50) >DroSim_CAF1 23119 120 + 1 CGGACUGCGCAUUUUGCAACUGGCUGACUCUUCCCAGCGCGCCAGCAAAAAAGUUACGCAUGCGCCAAGUGGCCCAAACUAUGGCCACCCGAUGGAAAGCCAGACAGCAGACCCACCAAC .((.((((((.....))..((((((.....((((..((((((.(((......)))..))..))))...((((((........)))))).....))))))))))...)))).))....... ( -40.00) >DroYak_CAF1 24911 99 + 1 CGGACUGCGCAUUGUGCAACUUG-UGGCUCUUCCCAGC--GCCAGCAAG-AAGUUACGCAUGCGCCAAGUGGCCAAAUCUAUGGCCACCCGAUGGAAAUCCAG----------------- (((...(((((((((((..((((-((((.((....)).--)))).))))-..).)))).))))))...(((((((......)))))))))).(((....))).----------------- ( -40.80) >consensus CGGACUGCGCAUUUUGCAACUGGCUGGCUCUUCCCAGC__GCCAGCAAAAAAGUUACGCAUGCGCCAAGUGGCCAAAACUAUGGCCACCCGAUGGAAAGCCAG_CAGCAGACC_ACC_AC (((...(((((...(((((((.((((((............)))))).....))))..))))))))...((((((........))))))))).(((....))).................. (-32.73 = -33.40 + 0.67)



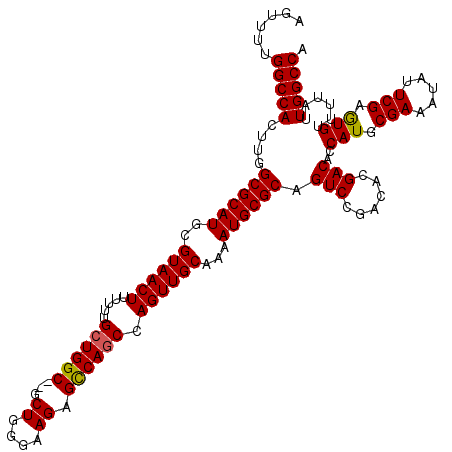
| Location | 14,465,233 – 14,465,351 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -52.57 |
| Consensus MFE | -41.66 |
| Energy contribution | -44.67 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14465233 118 - 27905053 GUCGGUCGGUCUGCUGGCUGGCUUUCCAUCGGGUGGCCAUAGUUUUGGCCACUUGGCGCAUGCGUAACUUUUUUGCUGGC--GCUGGGAAGAGCCAGCCAGUUGCAAAAUGCGCAGUCCG ......(((.(((((((((((((((((..((((((((((......))))))))))((((..(((.........)))..))--))..)).)))))))))))...((.....)))))).))) ( -61.50) >DroSim_CAF1 23119 120 - 1 GUUGGUGGGUCUGCUGUCUGGCUUUCCAUCGGGUGGCCAUAGUUUGGGCCACUUGGCGCAUGCGUAACUUUUUUGCUGGCGCGCUGGGAAGAGUCAGCCAGUUGCAAAAUGCGCAGUCCG .......((.((((((.((((((((((..(((((((((........)))))))))((((..(((.........)))..))))....)).)))))))).))...((.....)))))).)). ( -49.40) >DroYak_CAF1 24911 99 - 1 -----------------CUGGAUUUCCAUCGGGUGGCCAUAGAUUUGGCCACUUGGCGCAUGCGUAACUU-CUUGCUGGC--GCUGGGAAGAGCCA-CAAGUUGCACAAUGCGCAGUCCG -----------------..(((((.....((((((((((......))))))))))(((((((.((((((.-..((.((((--.((....)).))))-)))))))).).))))))))))). ( -46.80) >consensus GU_GGU_GGUCUGCUG_CUGGCUUUCCAUCGGGUGGCCAUAGUUUUGGCCACUUGGCGCAUGCGUAACUUUUUUGCUGGC__GCUGGGAAGAGCCAGCCAGUUGCAAAAUGCGCAGUCCG (((((((((....))))))))).......((((((((((......))))))))))((((((..((((((.....((((((...((....)).)))))).))))))...))))))...... (-41.66 = -44.67 + 3.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:34 2006