
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,464,996 – 14,465,153 |
| Length | 157 |
| Max. P | 0.781780 |

| Location | 14,464,996 – 14,465,113 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -40.27 |
| Consensus MFE | -32.87 |
| Energy contribution | -35.20 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14464996 117 + 27905053 CUUCAAAACCAUGCAAAUGUCAGAUGCCAUCUAUG-GUAGAACUUUCCUGUCUGGGCGAAUGUCGAUUGCCGUGAGUCGU--CUGGCCAGAAUUUGGAAAGUAUUCCCAGGCGGCAUCAA ......................((((((..((..(-(.((.(((((((..(((((.((...(.(((((......))))).--))).)))))....))))))).)).))))..)))))).. ( -37.10) >DroSim_CAF1 22887 120 + 1 CUUCAAAACCAUGCAAAUGUCAGAUGCCAUCUAUGGGUAGAACUUUCCUGUCUGGGCGAAUGUCGAUUGCCGUGCGGCGUCUCUGGCCAGAAUUAGGAAAGUAUUCCCAUGCGGCAUCAU ......................((((((...((((((.((.((((((((((((((.(((((((((.........)))))).)).).)))))..))))))))).)))))))).)))))).. ( -44.40) >DroYak_CAF1 24684 116 + 1 CUUCAAAACCAUGUAAAUGUCAGAUGCCAUCUAUA-GUAGAACUUUCCUGUCUGGGCGAAUGGCGAUUGCCGUGAGGCGU--CUGGCCAGAAUUUGGAAAGUAUUCCCAGGCGGCAUCA- ......................((((((.((((..-.))))(((((((..(((((((.....))(((.(((....)))))--)...)))))....)))))))..........)))))).- ( -39.30) >consensus CUUCAAAACCAUGCAAAUGUCAGAUGCCAUCUAUG_GUAGAACUUUCCUGUCUGGGCGAAUGUCGAUUGCCGUGAGGCGU__CUGGCCAGAAUUUGGAAAGUAUUCCCAGGCGGCAUCA_ ......................((((((.((((....))))(((((((..(((((.((.....)....(((....)))......).)))))....)))))))..........)))))).. (-32.87 = -35.20 + 2.33)

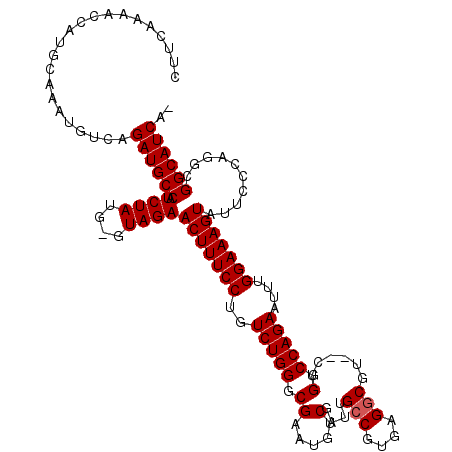

| Location | 14,465,035 – 14,465,153 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -41.03 |
| Consensus MFE | -29.31 |
| Energy contribution | -31.64 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14465035 118 + 27905053 AACUUUCCUGUCUGGGCGAAUGUCGAUUGCCGUGAGUCGU--CUGGCCAGAAUUUGGAAAGUAUUCCCAGGCGGCAUCAACAUCUCUAUGUGGCAUCAUUUGCUGUUGCCGCAACAUGCC .(((((((..(((((.((...(.(((((......))))).--))).)))))....)))))))........((((((.(.((((....))))((((.....))))).))))))........ ( -39.10) >DroSim_CAF1 22927 112 + 1 AACUUUCCUGUCUGGGCGAAUGUCGAUUGCCGUGCGGCGUCUCUGGCCAGAAUUAGGAAAGUAUUCCCAUGCGGCAUCAUCAUCUCUG--------CUGUUGCAGUUGCCGCAACAUGCC .((((((((((((((.(((((((((.........)))))).)).).)))))..))))))))).......(((((((.........(((--------(....)))).)))))))....... ( -42.30) >DroYak_CAF1 24723 110 + 1 AACUUUCCUGUCUGGGCGAAUGGCGAUUGCCGUGAGGCGU--CUGGCCAGAAUUUGGAAAGUAUUCCCAGGCGGCAUCA---UCUUGAAGUG----CAUUUGCAGCUGCCGCAACUUAC- .(((((((..(((((((.....))(((.(((....)))))--)...)))))....))))))).......((((((.((.---....))..((----(....))))))))).........- ( -41.70) >consensus AACUUUCCUGUCUGGGCGAAUGUCGAUUGCCGUGAGGCGU__CUGGCCAGAAUUUGGAAAGUAUUCCCAGGCGGCAUCA_CAUCUCUA_GUG____CAUUUGCAGUUGCCGCAACAUGCC .(((((((..(((((.((.....)....(((....)))......).)))))....)))))))........((((((..............................))))))........ (-29.31 = -31.64 + 2.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:31 2006