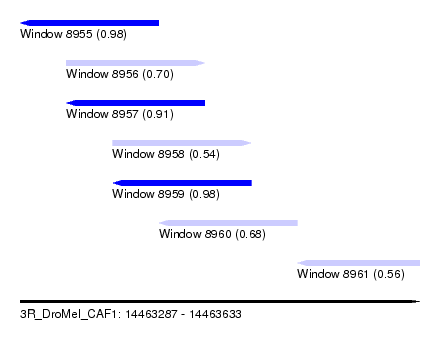
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,463,287 – 14,463,633 |
| Length | 346 |
| Max. P | 0.984501 |

| Location | 14,463,287 – 14,463,407 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -35.23 |
| Energy contribution | -35.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14463287 120 - 27905053 CGGCUGUCAGCCAACACAUUUGGCCAGGCCAACAAACAACGGCGGACCGCCCAGCUGACAAAUGCAAAUCUGUUGGCCAAGUCAACGGUCAACAAAGCGAUAAGUUUGGUAAAAAAAAAA .((((((..(((((.....)))))..((((((((......((((...))))..((........)).....))))))))......)))))).((.((((.....)))).)).......... ( -35.70) >DroSim_CAF1 21222 120 - 1 CGGCUGUCAGCCAACACAUUUGGCCAGGCCAACAAACAACGGCGGACCGCCCAGCUGACAAAUGCAAAUCUGUUGGCCAAGUCAAUGGUCAACAAAGCGAUAAGUUUGGUCAAAAAAAAA .(((.....)))......(((((((((((..........((((((.....)).)))).....(((.....(((((((((......)))))))))..)))....)))))))))))...... ( -37.60) >DroYak_CAF1 23100 118 - 1 CGGCUGUCAGCCAACACAUUUGGCCUGGCCAACAAACAACGGCGGACCGCCCAGCUGACAAAUGCAAAUCUGUUGGCCAAGUCAACGGUCAACAAAGCGAUAAGUUUGGUCGAAA--AAA .(((.....))).......(((((.(((((((((......((((...))))..((........)).....))))))))).)))))((..(((....((.....)))))..))...--... ( -37.90) >consensus CGGCUGUCAGCCAACACAUUUGGCCAGGCCAACAAACAACGGCGGACCGCCCAGCUGACAAAUGCAAAUCUGUUGGCCAAGUCAACGGUCAACAAAGCGAUAAGUUUGGUCAAAAAAAAA .((((....(((((.....)))))..))))..(((((..((((((.....)).)))).....(((.....((((((((........))))))))..)))....)))))............ (-35.23 = -35.23 + -0.00)

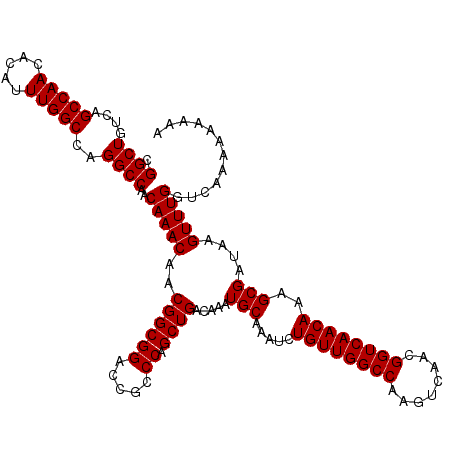
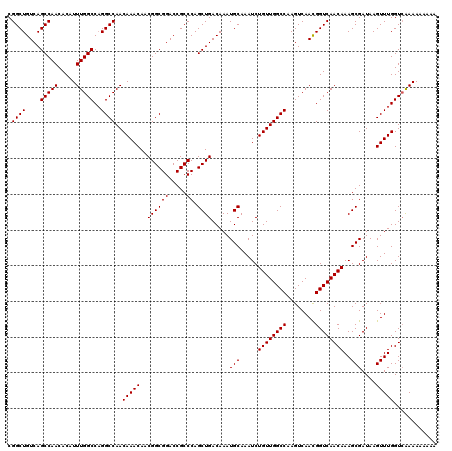
| Location | 14,463,327 – 14,463,447 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -38.70 |
| Energy contribution | -38.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14463327 120 + 27905053 UUGGCCAACAGAUUUGCAUUUGUCAGCUGGGCGGUCCGCCGUUGUUUGUUGGCCUGGCCAAAUGUGUUGGCUGACAGCCGCAUAAAUUGUUUUCUCAUGUGUGCCAAAUGUUUGACUUAC ..((((((((.((((((....((((((..(((((....)))))....))))))...).))))).))))))))((((...(((((...((......))..)))))....))))........ ( -38.70) >DroSim_CAF1 21262 120 + 1 UUGGCCAACAGAUUUGCAUUUGUCAGCUGGGCGGUCCGCCGUUGUUUGUUGGCCUGGCCAAAUGUGUUGGCUGACAGCCGCAUAAAUUGUUUUCUCAUGUGUGCCAAAUGUUUGACUUAC ..((((((((.((((((....((((((..(((((....)))))....))))))...).))))).))))))))((((...(((((...((......))..)))))....))))........ ( -38.70) >DroYak_CAF1 23138 120 + 1 UUGGCCAACAGAUUUGCAUUUGUCAGCUGGGCGGUCCGCCGUUGUUUGUUGGCCAGGCCAAAUGUGUUGGCUGACAGCCGCAUAAAUUGUUUUCUCAUGUGUGCCAAAUGUUUGACUUAC .((((((((((((..((........))..(((((....)))))))))))))))))((((((.....))))))((((...(((((...((......))..)))))....))))........ ( -39.10) >consensus UUGGCCAACAGAUUUGCAUUUGUCAGCUGGGCGGUCCGCCGUUGUUUGUUGGCCUGGCCAAAUGUGUUGGCUGACAGCCGCAUAAAUUGUUUUCUCAUGUGUGCCAAAUGUUUGACUUAC ..((((((((.((((((....((((((..(((((....)))))....))))))...).))))).))))))))((((...(((((...((......))..)))))....))))........ (-38.70 = -38.70 + -0.00)

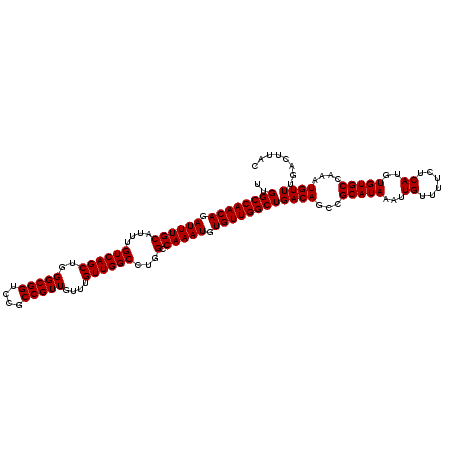
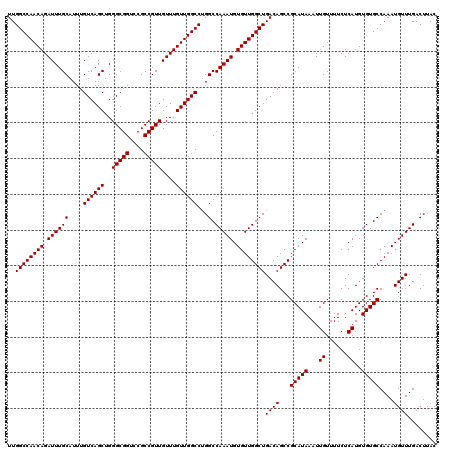
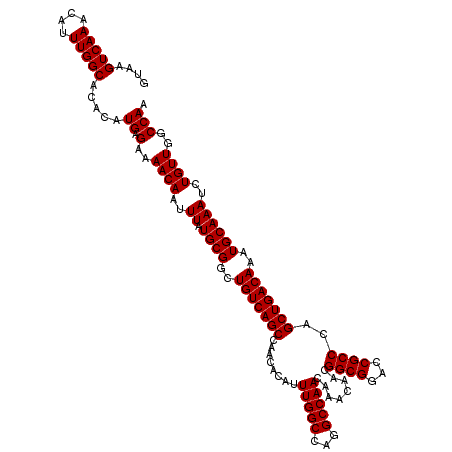
| Location | 14,463,327 – 14,463,447 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -32.70 |
| Energy contribution | -32.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14463327 120 - 27905053 GUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUGCGGCUGUCAGCCAACACAUUUGGCCAGGCCAACAAACAACGGCGGACCGCCCAGCUGACAAAUGCAAAUCUGUUGGCCAA ..............((((.((((((((.......)))))).(((.....)))........))))))((((((((......((((...))))..((........)).....)))))))).. ( -33.40) >DroSim_CAF1 21262 120 - 1 GUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUGCGGCUGUCAGCCAACACAUUUGGCCAGGCCAACAAACAACGGCGGACCGCCCAGCUGACAAAUGCAAAUCUGUUGGCCAA ..............((((.((((((((.......)))))).(((.....)))........))))))((((((((......((((...))))..((........)).....)))))))).. ( -33.40) >DroYak_CAF1 23138 120 - 1 GUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUGCGGCUGUCAGCCAACACAUUUGGCCUGGCCAACAAACAACGGCGGACCGCCCAGCUGACAAAUGCAAAUCUGUUGGCCAA ....((((((...((((((((((((((.......))))))....)))..)))))....)))))).(((((((((......((((...))))..((........)).....))))))))). ( -33.20) >consensus GUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUGCGGCUGUCAGCCAACACAUUUGGCCAGGCCAACAAACAACGGCGGACCGCCCAGCUGACAAAUGCAAAUCUGUUGGCCAA ....(((((....))))).....((.(..((((..((.((((..(((((((........(((((...)))))........((((...))))..)))))))..))))))..))))..))). (-32.70 = -32.70 + -0.00)

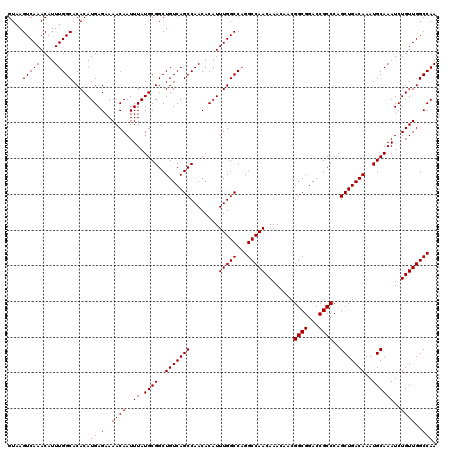
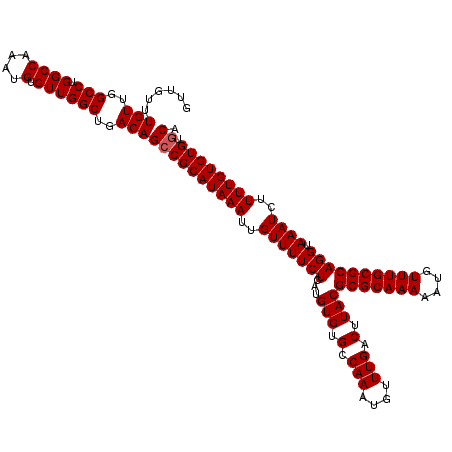
| Location | 14,463,367 – 14,463,487 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -32.83 |
| Energy contribution | -33.17 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14463367 120 + 27905053 GUUGUUUGUUGGCCUGGCCAAAUGUGUUGGCUGACAGCCGCAUAAAUUGUUUUCUCAUGUGUGCCAAAUGUUUGACUUACGCGCAAAAAUGUUUGCGCAGGUAAAUCUUUUGUGUGUGCA ......(((..(((.((((....).))))))..)))((((((((((..(((((((...(((.(.(((....))).).)))(((((((....)))))))))).))))..)))))))).)). ( -35.40) >DroSim_CAF1 21302 120 + 1 GUUGUUUGUUGGCCUGGCCAAAUGUGUUGGCUGACAGCCGCAUAAAUUGUUUUCUCAUGUGUGCCAAAUGUUUGACUUACGCGCAAAAAUGUUUGCGCAGGUAAAUCUUUUGUGUGUGCA ......(((..(((.((((....).))))))..)))((((((((((..(((((((...(((.(.(((....))).).)))(((((((....)))))))))).))))..)))))))).)). ( -35.40) >DroYak_CAF1 23178 120 + 1 GUUGUUUGUUGGCCAGGCCAAAUGUGUUGGCUGACAGCCGCAUAAAUUGUUUUCUCAUGUGUGCCAAAUGUUUGACUUACGCGCAAAAAUGUUUGCGCAGGUAAAUCUUUUGUGUGGACA .....((((..(((((..(....)..)))))..))))(((((((((..(((((((...(((.(.(((....))).).)))(((((((....)))))))))).))))..)))))))))... ( -37.30) >consensus GUUGUUUGUUGGCCUGGCCAAAUGUGUUGGCUGACAGCCGCAUAAAUUGUUUUCUCAUGUGUGCCAAAUGUUUGACUUACGCGCAAAAAUGUUUGCGCAGGUAAAUCUUUUGUGUGUGCA ......(((..(((.((((....).))))))..)))((((((((((..(((((((...(((.(.(((....))).).)))(((((((....)))))))))).))))..)))))))).)). (-32.83 = -33.17 + 0.33)

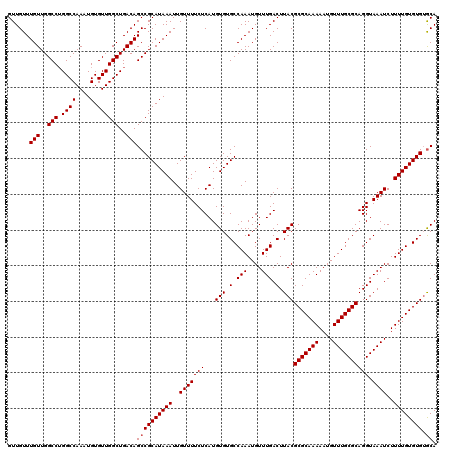
| Location | 14,463,367 – 14,463,487 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -30.10 |
| Energy contribution | -30.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14463367 120 - 27905053 UGCACACACAAAAGAUUUACCUGCGCAAACAUUUUUGCGCGUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUGCGGCUGUCAGCCAACACAUUUGGCCAGGCCAACAAACAAC .............(((((((..(((((((....))))))))))))))......(((((.(.((((((.......)))))).).(((...(((((.....)))))))))))))........ ( -31.60) >DroSim_CAF1 21302 120 - 1 UGCACACACAAAAGAUUUACCUGCGCAAACAUUUUUGCGCGUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUGCGGCUGUCAGCCAACACAUUUGGCCAGGCCAACAAACAAC .............(((((((..(((((((....))))))))))))))......(((((.(.((((((.......)))))).).(((...(((((.....)))))))))))))........ ( -31.60) >DroYak_CAF1 23178 120 - 1 UGUCCACACAAAAGAUUUACCUGCGCAAACAUUUUUGCGCGUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUGCGGCUGUCAGCCAACACAUUUGGCCUGGCCAACAAACAAC .............(((((((..(((((((....)))))))))))))).....((((((.(.((((((.......)))))).))).(((((((((.....)))).)))))...)))).... ( -31.00) >consensus UGCACACACAAAAGAUUUACCUGCGCAAACAUUUUUGCGCGUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUGCGGCUGUCAGCCAACACAUUUGGCCAGGCCAACAAACAAC .............(((((((..(((((((....))))))))))))))......(((((.(.((((((.......)))))).).......(((((.....)))))...)))))........ (-30.10 = -30.10 + -0.00)

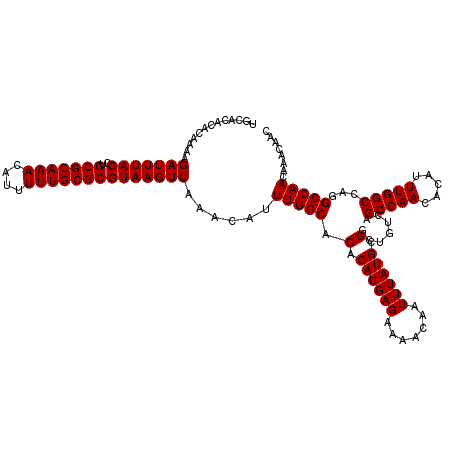
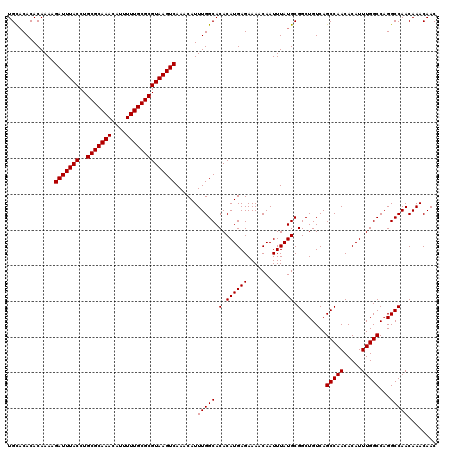
| Location | 14,463,407 – 14,463,527 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -20.96 |
| Energy contribution | -20.74 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14463407 120 - 27905053 GGAAAACAAGACAAAAAAUUUGACCCAAUCCGGUGCACGGUGCACACACAAAAGAUUUACCUGCGCAAACAUUUUUGCGCGUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUG (((...((((........))))......))).((((...(((....)))....(((((((..(((((((....)))))))))))))).........)))).((((((.......)))))) ( -24.70) >DroSim_CAF1 21342 113 - 1 GGAAAAAAAGACAAAAAAUUUGACCCAAU-------ACGGUGCACACACAAAAGAUUUACCUGCGCAAACAUUUUUGCGCGUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUG .............................-------...((((.((.......(((((((..(((((((....)))))))))))))).......)))))).((((((.......)))))) ( -21.64) >DroYak_CAF1 23218 112 - 1 GGAAAACAAGACAAA-AAUUUGACCCAAU-------CCGGUGUCCACACAAAAGAUUUACCUGCGCAAACAUUUUUGCGCGUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUG (((........(((.-...)))......)-------)).(((((((.......(((((((..(((((((....)))))))))))))).......))).)))).................. ( -22.88) >consensus GGAAAACAAGACAAAAAAUUUGACCCAAU_______ACGGUGCACACACAAAAGAUUUACCUGCGCAAACAUUUUUGCGCGUAAGUCAAACAUUUGGCACACAUGAGAAAACAAUUUAUG .......................................((((.((.......(((((((..(((((((....)))))))))))))).......)))))).((((((.......)))))) (-20.96 = -20.74 + -0.22)

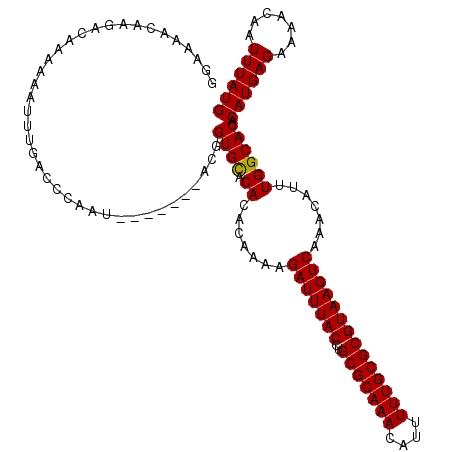
| Location | 14,463,527 – 14,463,633 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 91.72 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -22.15 |
| Energy contribution | -21.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14463527 106 - 27905053 AAUUGCCAGGUGGAGGAAAUAACUUUGCACG----------CAAGCAUUUCGCAUUUUUGUGUAUAAAUUAAUUGAUUUAUAUGCAUGGCUAGCAACAAAAUACAAAUGCUCUUGC ..((((...(..(((.......)))..)..)----------)))(((....((((((.((((((((((((....))))))))))))((........))......))))))...))) ( -24.60) >DroSim_CAF1 21455 106 - 1 AAUUGCCAGGUGUAGGAAAUAACUUCGCACG----------CAAGCAUUUCGCAUUUUUGUGUAUAAAUUAAUUGAUUUAUAUGCAUGGCUAGCAACAAAAUACAAAUGCUCUUGC ..((((...(((.((.......)).)))..)----------)))(((....((((((.((((((((((((....))))))))))))((........))......))))))...))) ( -22.20) >DroYak_CAF1 23330 116 - 1 AAUUGCCAGGUGGAGGAAAUAACUCCGCACGCAAUGAUCCGCAAGCAUUUCGCAUUUUUGUGUAUAAAUUAAUUGAUUUAUAUGCAUGGCUAGCAACAAAAUACAAAUGUUCUUGC .(((((...((((((.......))))))..))))).....((((((((((.(...(((((((((((((((....)))))))))((.......)).))))))..))))))..))))) ( -30.20) >consensus AAUUGCCAGGUGGAGGAAAUAACUUCGCACG__________CAAGCAUUUCGCAUUUUUGUGUAUAAAUUAAUUGAUUUAUAUGCAUGGCUAGCAACAAAAUACAAAUGCUCUUGC ....((((.((((((.......))))))................((.....)).....((((((((((((....)))))))))))))))).((((............))))..... (-22.15 = -21.93 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:27 2006