| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,047,719 – 2,047,819 |
| Length | 100 |
| Max. P | 0.792168 |

| Location | 2,047,719 – 2,047,819 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 73.51 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -15.57 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2047719 100 + 27905053 AUCGCAC-GUUUCUGCUACAGUUUUGAUUUCAAGUCAACCAGCUGAGUGACGGCCAUCGCUCAGCGGGCAGGCUACAGUAUUUCAGUAUCGCAACACCAGA------ ...((..-((.(((((.......(((((.....)))))((.(((((((((......))))))))))))))))..)).((((....)))).)).........------ ( -27.70) >DroPse_CAF1 167789 94 + 1 --CAAAC-AAUUUUGCUAGAGUUUUGAUUUCAAGUCAACCAGCUGAGUGACGGCCAUCGCUCAGCAGGCAG-CCGCAGCAU--------CAGAAAU-CAGACUCUGA --.....-........((((((((.((((((.......((.(((((((((......))))))))).))..(-(....))..--------..)))))-))))))))). ( -32.30) >DroSec_CAF1 190815 98 + 1 --CGCAC-GUUUCUGCUACAGUUUUGAUUUCAAGUCAACCAGCUUAGUGACGGCCAUCGCUCAGCGGGCAAGCUCCAGUAUUUCAGUAUUGCAACACCAGA------ --.((.(-(((...((((.(((((((((.....)))))..))))))))))))(((..(((...))))))..))..((((((....))))))..........------ ( -20.10) >DroSim_CAF1 195703 98 + 1 --CGCAC-GUUUCUGCUACAGUUUUGAUUUCAAGUCAACCAGCUGAGUGACGGCCAUCGCUCAGCGGGCAAGCUCCAGUAUUUCAGUAUCGCAACACCAGA------ --.((((-.....((((..(((((((((.....)))).((.(((((((((......))))))))).)).)))))..)))).....))...)).........------ ( -24.30) >DroAna_CAF1 167873 91 + 1 AUUGCCCAGUUUCUUCCACAGUUUGGAUUUCA-GUGAACCAUCUGAGUGACGGCCAUCGCUCAGCGGGCAGGCUUC--------GGUAUCGAAGCA-CAGA------ .((((((.((((((((((.....))))....)-).))))...((((((((......)))))))).))))))(((((--------(....)))))).-....------ ( -32.40) >DroPer_CAF1 169331 94 + 1 --NNNNN-NNNNNNNNNACAGUUUUGAUUUCAAGUCAACCAGCUGAGUGACGGCCAUCGCUCAGCAGGCAG-CCGCAGCAU--------CAGAAAU-CAGACUCUGA --.....-..........(((.(((((((((.......((.(((((((((......))))))))).))..(-(....))..--------..)))))-))))..))). ( -29.60) >consensus __CGCAC_GUUUCUGCUACAGUUUUGAUUUCAAGUCAACCAGCUGAGUGACGGCCAUCGCUCAGCGGGCAGGCUCCAGUAU___AGUAUCGCAACA_CAGA______ .......................(((((.....)))))((.(((((((((......))))))))).))....................................... (-15.57 = -16.23 + 0.67)


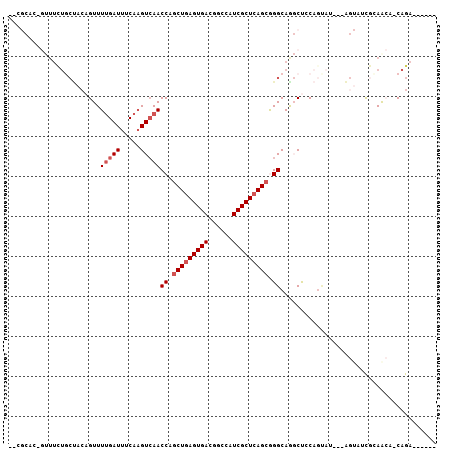
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:49 2006