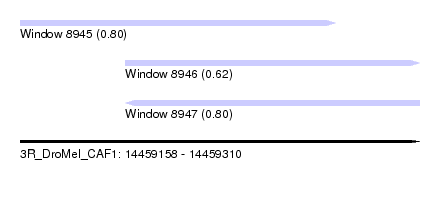
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,459,158 – 14,459,310 |
| Length | 152 |
| Max. P | 0.797535 |

| Location | 14,459,158 – 14,459,278 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -38.84 |
| Consensus MFE | -34.95 |
| Energy contribution | -35.45 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14459158 120 + 27905053 UAAUGCCCUGCGUCAGGGUAUAAAGAGUAGCCAUAAAAAGAUUUAAUUUUUUCGGUGUUUUUUGGACAACAACGGCGGAUGGGAGUGCGUCGUCAUGGGGCACACCCCUUUGCCACGUCA ..((((((((...))))))))...((((.((((.(((((.((..((....))..)).)))))))).).))...((((((.(((.((((.((.....)).)))).))).))))))...)). ( -39.30) >DroSec_CAF1 16509 119 + 1 CAAUGCCCUGCUUCAGGGUAUAAAGAGUAGCCAUAAAAAGAUUUAAUU-UUUCGGUGUUUUUUGGACACCAACGGCGGAUGGGAGUGCGUCGUCAUGGGGCACACCCCUUUGCCACGUCA ..((((((((...))))))))..................(((......-....((((((.....))))))...((((((.(((.((((.((.....)).)))).))).))))))..))). ( -42.00) >DroSim_CAF1 17124 119 + 1 CAAUGCCCUGCGUCAGGGUAUAAAGAGUAGCCAUAAAAAGAUUUAAUU-UUUCGGUGUUUUUUGGACACCAACGGCGGAUGGGAGUGCGUCGUCAUGGGGCACACCCCUUUGCCACGUCA ..((((((((...))))))))..................(((......-....((((((.....))))))...((((((.(((.((((.((.....)).)))).))).))))))..))). ( -42.00) >DroYak_CAF1 19038 117 + 1 UAUUGCCCUGCGUCAGAGUA--AAAAAUAGCCAUAAAAAGAUUUAAUU-UUUCUGUGUUUUUUGGGGACCAACGGCGGAUGGGAGUGCGUCGUCAUGGGGCACACCCCUUUGCCACGUCA .....(((.(((.(((((..--..........................-.)))))))).....)))(((....((((((.(((.((((.((.....)).)))).))).))))))..))). ( -32.05) >consensus CAAUGCCCUGCGUCAGGGUAUAAAGAGUAGCCAUAAAAAGAUUUAAUU_UUUCGGUGUUUUUUGGACACCAACGGCGGAUGGGAGUGCGUCGUCAUGGGGCACACCCCUUUGCCACGUCA ..((((((((...)))))))).........(((.(((((.((..((....))..)).))))))))........((((((.(((.((((.((.....)).)))).))).))))))...... (-34.95 = -35.45 + 0.50)


| Location | 14,459,198 – 14,459,310 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.45 |
| Mean single sequence MFE | -40.35 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.57 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14459198 112 + 27905053 AUUUAAUUUUUUCGGUGUUUUUUGGACAACAACGGCGGAUGGGAGUGCGUCGUCAUGGGGCACACCCCUUUGCCACGUCAGGCGUUGUGGCUAGCUGGUUGUC--------UGUCUGUCC .......................((((((((((((((((.(((.((((.((.....)).)))).))).)))((((((........))))))..))).))))).--------))))).... ( -36.30) >DroSec_CAF1 16549 119 + 1 AUUUAAUU-UUUCGGUGUUUUUUGGACACCAACGGCGGAUGGGAGUGCGUCGUCAUGGGGCACACCCCUUUGCCACGUCAGGCGUUGUGGCUAGCUGGUUGUCUGUCUGUCUGACUGUCC ........-....((((((.....))))))...((((((.(((.((((.((.....)).)))).))).))))))..((((((((....(((..((.....))..)))))))))))..... ( -44.60) >DroSim_CAF1 17164 116 + 1 AUUUAAUU-UUUCGGUGUUUUUUGGACACCAACGGCGGAUGGGAGUGCGUCGUCAUGGGGCACACCCCUUUGCCACGUCAGGCGUUGUGGCUAGCUGG---UCUGUCUGUCUGCCUGUCC ........-....((((((.....))))))...((((((.(((.((((.((.....)).)))).))).))))))..(.((((((....(((..((...---...))..))))))))).). ( -43.20) >DroYak_CAF1 19076 103 + 1 AUUUAAUU-UUUCUGUGUUUUUUGGGGACCAACGGCGGAUGGGAGUGCGUCGUCAUGGGGCACACCCCUUUGCCACGUCAGGCGUUGUGGCUAGUUGGUUGUC----------------U ........-..............((.(((((((((((((.(((.((((.((.....)).)))).))).))))))......(((......))).))))))).))----------------. ( -37.30) >consensus AUUUAAUU_UUUCGGUGUUUUUUGGACACCAACGGCGGAUGGGAGUGCGUCGUCAUGGGGCACACCCCUUUGCCACGUCAGGCGUUGUGGCUAGCUGGUUGUC________UG_CUGUCC .............((((((.....))))))..(((((((.(((.((((.((.....)).)))).))).)))((((((........))))))..))))....................... (-31.20 = -31.57 + 0.37)

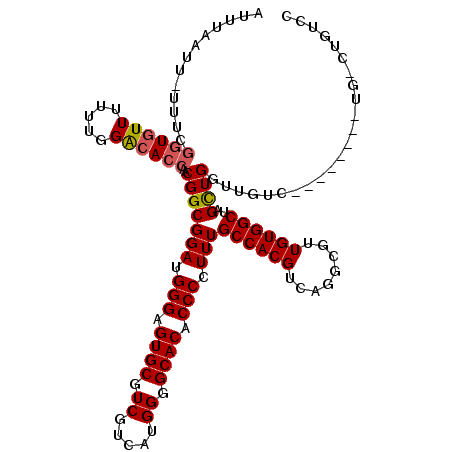
| Location | 14,459,198 – 14,459,310 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.45 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -24.81 |
| Energy contribution | -26.12 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14459198 112 - 27905053 GGACAGACA--------GACAACCAGCUAGCCACAACGCCUGACGUGGCAAAGGGGUGUGCCCCAUGACGACGCACUCCCAUCCGCCGUUGUUGUCCAAAAAACACCGAAAAAAUUAAAU (((((....--------.(((((..((..(((((..........)))))...(((((((((........).))))).)))....)).))))))))))....................... ( -31.20) >DroSec_CAF1 16549 119 - 1 GGACAGUCAGACAGACAGACAACCAGCUAGCCACAACGCCUGACGUGGCAAAGGGGUGUGCCCCAUGACGACGCACUCCCAUCCGCCGUUGGUGUCCAAAAAACACCGAAA-AAUUAAAU ((((.(((.....))).....((((((..(((((..........)))))...(((((((((........).))))).))).......))))))))))..............-........ ( -36.20) >DroSim_CAF1 17164 116 - 1 GGACAGGCAGACAGACAGA---CCAGCUAGCCACAACGCCUGACGUGGCAAAGGGGUGUGCCCCAUGACGACGCACUCCCAUCCGCCGUUGGUGUCCAAAAAACACCGAAA-AAUUAAAU ((((.(.....)......(---(((((..(((((..........)))))...(((((((((........).))))).))).......))))))))))..............-........ ( -32.60) >DroYak_CAF1 19076 103 - 1 A----------------GACAACCAACUAGCCACAACGCCUGACGUGGCAAAGGGGUGUGCCCCAUGACGACGCACUCCCAUCCGCCGUUGGUCCCCAAAAAACACAGAAA-AAUUAAAU .----------------....((((((..(((((..........)))))...(((((((((........).))))).))).......))))))..................-........ ( -26.50) >consensus GGACAG_CA________GACAACCAGCUAGCCACAACGCCUGACGUGGCAAAGGGGUGUGCCCCAUGACGACGCACUCCCAUCCGCCGUUGGUGUCCAAAAAACACCGAAA_AAUUAAAU (((((.................(((((..(((((..........)))))...(((((((((........).))))).))).......))))))))))....................... (-24.81 = -26.12 + 1.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:14 2006