
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,045,971 – 2,046,091 |
| Length | 120 |
| Max. P | 0.637469 |

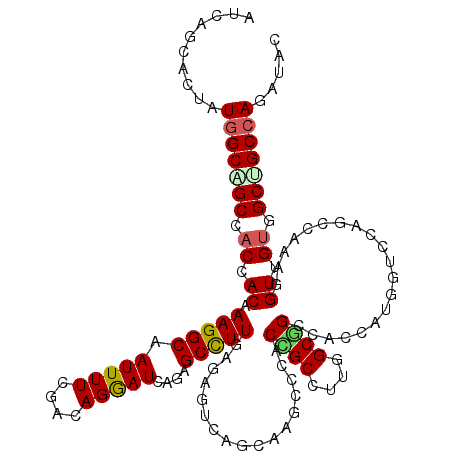
| Location | 2,045,971 – 2,046,091 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.44 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -23.39 |
| Energy contribution | -23.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2045971 120 + 27905053 AUCAACACUAUUGCGGCUACCACGAAGGCAAUUUUGGACAGGAUCAGAGCCUUGAGAGUCAGGAGCCCCACCGCCUUUGCGGCCACCAUCGUCCACCCGAAUGUUGUGGCCGCCAGGUAC ...........((.((((.((.(((((....)))))(((....((((....))))..))).)))))).))..((((..(((((((((((((......)).)))..)))))))).)))).. ( -37.30) >DroVir_CAF1 237945 120 + 1 AUCAGCACAAUGGCGGCCACGACAAAGGCUAUUUUGGAAAGGAUGAGCGCCUUGAGGGUCAGCAGGCCCACCGCCUUGGCGGCCACCAUGGUCCAGCCAAAGGUUGUCGCGGCCAGAUAC (((.((......))((((.((((((((((((((((....))))))...)))))..(((((....)))))...((((((((((((.....)).)).))).)))))))))).)))).))).. ( -50.70) >DroGri_CAF1 190365 120 + 1 AUCAGCACAAUGGCUGCAACAACAAAGGCAAUUUUAGACAGAAUGAGCGCCUUAAGAGUGAGUAAGCCAACCGCCUUGGCGGCCACCAUCGUCCAUCCGAAGGUUGUGGCUGCCAGAUAC (((.((....(((((((.((....(((((.(((((....)))))....)))))....))..)).)))))...))..((((((((((.(((.((.....)).))).))))))))))))).. ( -37.60) >DroWil_CAF1 189913 120 + 1 AUCAAGACUAUGGCAGCCACCACAAAGGCAAUUUUCGAUAGAAUCAAAGCUUUUAGAGUCAACAAUCCCACAGCUUUUGCUGCCACCAUUGUCCAACCGAAUGUUGUGGCUGCCAGAUAC ..........((((((((((.((((((((....(((....))).....)))))).......(((((....((((....)))).....)))))..........)).))))))))))..... ( -34.80) >DroMoj_CAF1 194002 120 + 1 AUCAGCACGAUGGCUGCAACGACAAAGGCAAUUUUCGACAGGAUUAGAGCCUUGAGGGUCAGCAAACCCACCGCCUUGGCAGCCACCAUGGUCCAGCCAAAGGUUGAGGCAGCCAGAUAC ..........(((((((..((((.(((((.........((((........)))).((((......))))...)))))(((.(((.....)))...)))....))))..)))))))..... ( -41.40) >DroAna_CAF1 166149 120 + 1 AUGAGCACUAUGGCAGCCACCACAAAGGCUAUUUUUGACAGGAUCAAAGCCUUCAAGGUAAGCAGGCCCACUGCCUUCGCAGCCACCAUGGUCCAGCCAAAGGUCGUGGCUGCCAAGUAC ......(((.((((((((((.((...((((..((((((.(((.......)))))))))...((((((.....))))..))))))....(((.....)))...)).))))))))))))).. ( -44.00) >consensus AUCAGCACUAUGGCAGCCACCACAAAGGCAAUUUUCGACAGGAUCAGAGCCUUGAGAGUCAGCAAGCCCACCGCCUUGGCGGCCACCAUGGUCCAGCCAAAGGUUGUGGCUGCCAGAUAC ..........(((((((.((.((.(((((.(((((....)))))....))))).................((((....))))....................)).)).)))))))..... (-23.39 = -23.45 + 0.06)

| Location | 2,045,971 – 2,046,091 |
|---|---|
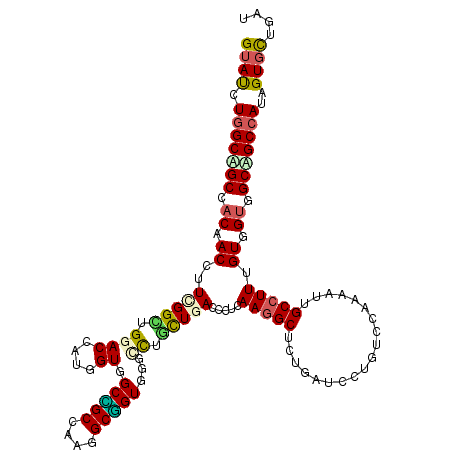
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.44 |
| Mean single sequence MFE | -43.79 |
| Consensus MFE | -27.91 |
| Energy contribution | -27.58 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2045971 120 - 27905053 GUACCUGGCGGCCACAACAUUCGGGUGGACGAUGGUGGCCGCAAAGGCGGUGGGGCUCCUGACUCUCAAGGCUCUGAUCCUGUCCAAAAUUGCCUUCGUGGUAGCCGCAAUAGUGUUGAU ..(((((((((((((.....(((......)))..))))))))....(((((((((((..((.....)).))))))...............((((.....)))))))))..))).)).... ( -41.50) >DroVir_CAF1 237945 120 - 1 GUAUCUGGCCGCGACAACCUUUGGCUGGACCAUGGUGGCCGCCAAGGCGGUGGGCCUGCUGACCCUCAAGGCGCUCAUCCUUUCCAAAAUAGCCUUUGUCGUGGCCGCCAUUGUGCUGAU ((((.((((((((((((.....(((((((....((((..((((..((.(((.((....)).))).))..))))..))))...))).....)))).)))))))))))).....)))).... ( -47.10) >DroGri_CAF1 190365 120 - 1 GUAUCUGGCAGCCACAACCUUCGGAUGGACGAUGGUGGCCGCCAAGGCGGUUGGCUUACUCACUCUUAAGGCGCUCAUUCUGUCUAAAAUUGCCUUUGUUGUUGCAGCCAUUGUGCUGAU ((((.((((.((.((((((....).(((((((((((((((((....))))))(.((((........)))).))).))))..)))))...........))))).)).))))..)))).... ( -36.50) >DroWil_CAF1 189913 120 - 1 GUAUCUGGCAGCCACAACAUUCGGUUGGACAAUGGUGGCAGCAAAAGCUGUGGGAUUGUUGACUCUAAAAGCUUUGAUUCUAUCGAAAAUUGCCUUUGUGGUGGCUGCCAUAGUCUUGAU .....((((((((((.(((...(((.......((((((.((..((((((...(((........)))...))))))..))))))))......)))..))).)))))))))).......... ( -39.42) >DroMoj_CAF1 194002 120 - 1 GUAUCUGGCUGCCUCAACCUUUGGCUGGACCAUGGUGGCUGCCAAGGCGGUGGGUUUGCUGACCCUCAAGGCUCUAAUCCUGUCGAAAAUUGCCUUUGUCGUUGCAGCCAUCGUGCUGAU ...((..(((............)))..)).((((((((((((...(((((.(((((....))))).)(((((.....((.....)).....)))))))))...))))))))))))..... ( -43.80) >DroAna_CAF1 166149 120 - 1 GUACUUGGCAGCCACGACCUUUGGCUGGACCAUGGUGGCUGCGAAGGCAGUGGGCCUGCUUACCUUGAAGGCUUUGAUCCUGUCAAAAAUAGCCUUUGUGGUGGCUGCCAUAGUGCUCAU (((((((((((((((.((....(((.((.((......(((((....))))).)))).)))......((((((((((((...)))))....))))))))).)))))))))).))))).... ( -54.40) >consensus GUAUCUGGCAGCCACAACCUUCGGCUGGACCAUGGUGGCCGCCAAGGCGGUGGGCCUGCUGACCCUCAAGGCUCUGAUCCUGUCCAAAAUUGCCUUUGUGGUGGCAGCCAUAGUGCUGAU ((((.(((((((.((.((..(((((.((((....)).(((((....)))))...)).))))).....(((((...................))))).)).)).)))))))..)))).... (-27.91 = -27.58 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:48 2006