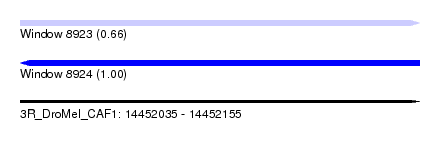
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,452,035 – 14,452,155 |
| Length | 120 |
| Max. P | 0.996916 |

| Location | 14,452,035 – 14,452,155 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -25.28 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
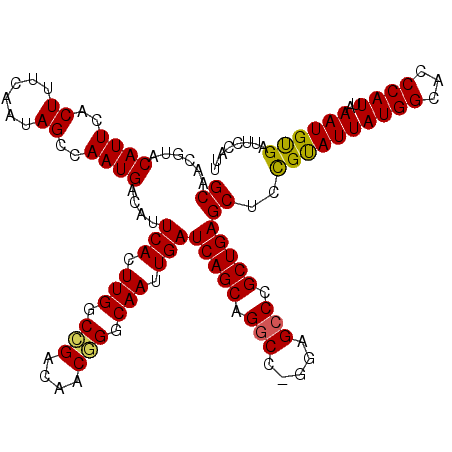
>3R_DroMel_CAF1 14452035 120 + 27905053 GCAAUGUACAUUCACUUUCAAUAGCCAAUGACAUUCACUUGGCCGACAACGGGCAAUUGAUCAGCAGGCCCAGAGCCUGCUGAGCUCCGUAUUAUGGCACCCAUUAAAUGUGAUUCCAUU ..((((.....((((.((.(((.((((...((..(((.(((.(((....))).))).)))(((((((((.....))))))))).....))....))))....))).)).))))...)))) ( -35.10) >DroSec_CAF1 9232 119 + 1 GCAACGUACAUUCACUUUCAAUAGCCAAUGACAUUCACUUGGCCGACAACGGGCAAUUGAUCAGCAGGCC-GUAGCCCGCUGAGCUCCGCAUUAUGGCACCCAUUAAAUGUGAUUCCAUU ((...((.((((..((......))..))))))..(((.(((.(((....))).))).)))(((((.(((.-...))).)))))))..((((((((((...))))..))))))........ ( -32.10) >DroSim_CAF1 9811 119 + 1 GCAACGUACAUUCACUUUCAAUAGCCAAUGACAUUCACUUGGCCGACAACGGGCAAUUGAUCAGCAGGCC-GUAGCCCGCUGAGCUCCGUAUUAUGGCACCCAUUAAAUGUGAUUCCAUU ...........((((.((.(((.((((...((..(((.(((.(((....))).))).)))(((((.(((.-...))).))))).....))....))))....))).)).))))....... ( -30.20) >DroEre_CAF1 9019 119 + 1 GCAACGAACAUUCACUUUCAAUAGCCAAUGACAUUCACUUGCCAGACAACUGGCAAUUGAUCAGCAGGCC-GGCGCCCGCUGAGCUCUGUAUUAUGGCACCCAUUAAAUGCGAUUCCAAU (((..(((........)))....((((.(((...(((.(((((((....))))))).))))))(((((((-(((....)))).)).))))....))))..........)))......... ( -35.80) >DroYak_CAF1 10366 119 + 1 GCAACGAACAUUCACUUUCAAUAGCCAAUGACAUUCACUUGGCCGACAACUGGCAAUUGAUCAGCAGCCC-GGAGCCCGCUGAGCUCUGUAUUAUGGUACCCAUUAAAUGCGAUUCCAUU ((...............(((((.((((.((.....)).(((.....))).)))).)))))(((((.((..-...))..)))))))..((((((((((...))))..))))))........ ( -23.00) >consensus GCAACGUACAUUCACUUUCAAUAGCCAAUGACAUUCACUUGGCCGACAACGGGCAAUUGAUCAGCAGGCC_GGAGCCCGCUGAGCUCCGUAUUAUGGCACCCAUUAAAUGUGAUUCCAUU ((......((((..((......))..))))....(((.(((.(((....))).))).)))(((((.(((.....))).)))))))..((((((((((...))))..))))))........ (-25.28 = -24.80 + -0.48)

| Location | 14,452,035 – 14,452,155 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -35.24 |
| Energy contribution | -34.60 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
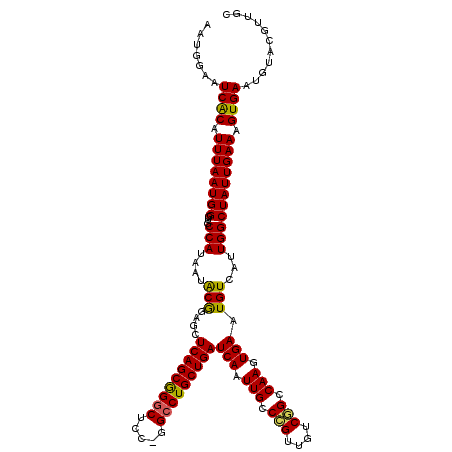
>3R_DroMel_CAF1 14452035 120 - 27905053 AAUGGAAUCACAUUUAAUGGGUGCCAUAAUACGGAGCUCAGCAGGCUCUGGGCCUGCUGAUCAAUUGCCCGUUGUCGGCCAAGUGAAUGUCAUUGGCUAUUGAAAGUGAAUGUACAUUGC ((((...((((...((((((((.((.......)).(.(((((((((.....))))))))).)....))))))))..((((((.((.....)))))))).......)))).....)))).. ( -41.60) >DroSec_CAF1 9232 119 - 1 AAUGGAAUCACAUUUAAUGGGUGCCAUAAUGCGGAGCUCAGCGGGCUAC-GGCCUGCUGAUCAAUUGCCCGUUGUCGGCCAAGUGAAUGUCAUUGGCUAUUGAAAGUGAAUGUACGUUGC ((((...((((...((((((((((......))...(.(((((((((...-.))))))))).)....))))))))..((((((.((.....)))))))).......)))).....)))).. ( -43.30) >DroSim_CAF1 9811 119 - 1 AAUGGAAUCACAUUUAAUGGGUGCCAUAAUACGGAGCUCAGCGGGCUAC-GGCCUGCUGAUCAAUUGCCCGUUGUCGGCCAAGUGAAUGUCAUUGGCUAUUGAAAGUGAAUGUACGUUGC ((((...((((...((((((((.((.......)).(.(((((((((...-.))))))))).)....))))))))..((((((.((.....)))))))).......)))).....)))).. ( -43.00) >DroEre_CAF1 9019 119 - 1 AUUGGAAUCGCAUUUAAUGGGUGCCAUAAUACAGAGCUCAGCGGGCGCC-GGCCUGCUGAUCAAUUGCCAGUUGUCUGGCAAGUGAAUGUCAUUGGCUAUUGAAAGUGAAUGUUCGUUGC ...(((.((((.(((((((...((((....(((....(((((((((...-.)))))))))(((.(((((((....))))))).))).)))...))))))))))).))))...)))..... ( -44.50) >DroYak_CAF1 10366 119 - 1 AAUGGAAUCGCAUUUAAUGGGUACCAUAAUACAGAGCUCAGCGGGCUCC-GGGCUGCUGAUCAAUUGCCAGUUGUCGGCCAAGUGAAUGUCAUUGGCUAUUGAAAGUGAAUGUUCGUUGC .((((.(((.(((...)))))).))))......(((((((((((.(...-.).)))))))(((((.((((((.(.((..(....)..)).))))))).)))))........))))..... ( -34.10) >consensus AAUGGAAUCACAUUUAAUGGGUGCCAUAAUACGGAGCUCAGCGGGCUCC_GGCCUGCUGAUCAAUUGCCCGUUGUCGGCCAAGUGAAUGUCAUUGGCUAUUGAAAGUGAAUGUACGUUGC .......((((.((((((((...(((....(((....(((((((((.....)))))))))(((.(((.(((....))).))).))).)))...))))))))))).))))........... (-35.24 = -34.60 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:52 2006