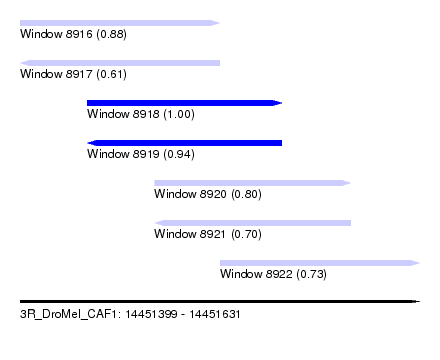
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,451,399 – 14,451,631 |
| Length | 232 |
| Max. P | 0.998530 |

| Location | 14,451,399 – 14,451,515 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.22 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -19.84 |
| Energy contribution | -19.48 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14451399 116 + 27905053 CCUUACUCCCCGAUCCUUCCUUGCC-ACAAAACCGUCCACAACAUUCCGAUGCACC-CGCAACAGCAUUGUGGUUGCCUCAUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAAC-- ...............((((......-........((.....)).....((.(((.(-(((((.....)))))).))).)).....))))((((((.((....)).)))))).......-- ( -24.80) >DroSec_CAF1 8609 119 + 1 ACUUACUCCCCGAUCCUUCCUUGCC-ACAAAACCAUCCACAACAUUCCGAUGCACCCCGCAACAGCAUUGUGGUUGCCUCAUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAACAA ..........((..........(((-((((...((((...........))))......((....)).)))))))..........))..(((((((.((....)).)))))))........ ( -23.05) >DroSim_CAF1 9203 118 + 1 ACAUACUCCCCGAUCCUUCCUUGCC-ACAAAACCGUCCACAACAUUCCGAUGCACC-CGCAACAGCAUUGUGGUUGCCUCAUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAACAA ...............((((......-........((.....)).....((.(((.(-(((((.....)))))).))).)).....))))((((((.((....)).))))))......... ( -24.80) >DroEre_CAF1 8407 116 + 1 CCUUACUCCGCAAUCCUUCCUUGCCCACAA-ACCGUCCACAACAUUCCGAAGCCCC-CGCAACUGCAUUGUGGUUGCCUCAUUACUACGGGGAAAUGCUCACGCUUUUCCCGGCCGAC-- .........((((.......))))......-..((.((..........((.((..(-(((((.....))))))..)).))........(((((((.((....))))))))))).))..-- ( -26.40) >DroYak_CAF1 9717 114 + 1 CUCUACUCCGCAAUCCUUCCUUGCCCACAAAACCAUCCACAACAUUCCGAUGCCCC-CGCAACUGCAUUGUGGUUGCCUCAUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCC----- .........(((...((((..............((((...........))))....-.(((((..(...)..)))))........))))((((((.((....)).))))))))).----- ( -25.00) >consensus CCUUACUCCCCGAUCCUUCCUUGCC_ACAAAACCGUCCACAACAUUCCGAUGCACC_CGCAACAGCAUUGUGGUUGCCUCAUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAAC__ ......................((.........((.((((((....(.(.(((.....))).).)..)))))).))...........((((((((.((....))))))))))))...... (-19.84 = -19.48 + -0.36)

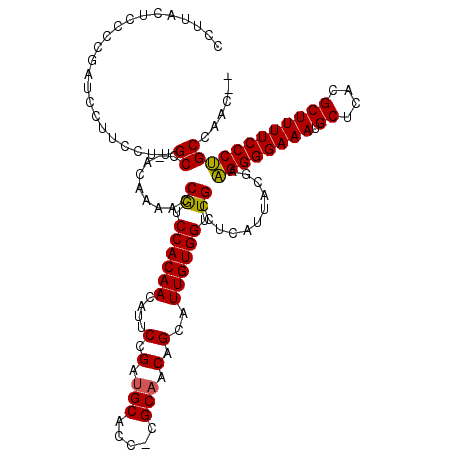
| Location | 14,451,399 – 14,451,515 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.22 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -31.26 |
| Energy contribution | -32.06 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14451399 116 - 27905053 --GUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAUGAGGCAACCACAAUGCUGUUGCG-GGUGCAUCGGAAUGUUGUGGACGGUUUUGU-GGCAAGGAAGGAUCGGGGAGUAAGG --....((.((((((.((....)).))))))((((((.(..((((..(((((((.(((.(((.-...))).)))...)))))))...))))..)-.))...)))).........)).... ( -36.80) >DroSec_CAF1 8609 119 - 1 UUGUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAUGAGGCAACCACAAUGCUGUUGCGGGGUGCAUCGGAAUGUUGUGGAUGGUUUUGU-GGCAAGGAAGGAUCGGGGAGUAAGU ((.(((((.((((((.((....)).))))))((((((.(..((((..(((((((.(((.(((.....))).)))...)))))))...))))..)-.))...))))).)))).))...... ( -37.60) >DroSim_CAF1 9203 118 - 1 UUGUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAUGAGGCAACCACAAUGCUGUUGCG-GGUGCAUCGGAAUGUUGUGGACGGUUUUGU-GGCAAGGAAGGAUCGGGGAGUAUGU ((.(((((.((((((.((....)).))))))((((((.(..((((..(((((((.(((.(((.-...))).)))...)))))))...))))..)-.))...))))).)))).))...... ( -37.60) >DroEre_CAF1 8407 116 - 1 --GUCGGCCGGGAAAAGCGUGAGCAUUUCCCCGUAGUAAUGAGGCAACCACAAUGCAGUUGCG-GGGGCUUCGGAAUGUUGUGGACGGU-UUGUGGGCAAGGAAGGAUUGCGGAGUAAGG --(((.((.((((((.((....)).))))))........((((((..((.((((...)))).)-)..)))))).......)).)))...-......((((.......))))......... ( -37.10) >DroYak_CAF1 9717 114 - 1 -----GGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAUGAGGCAACCACAAUGCAGUUGCG-GGGGCAUCGGAAUGUUGUGGAUGGUUUUGUGGGCAAGGAAGGAUUGCGGAGUAGAG -----....((((((.((....)).))))))(((((((((....((.(((((((.(...(((.-...)))...)...))))))).)).(((((....)))))....)))))))))..... ( -37.40) >consensus __GUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAUGAGGCAACCACAAUGCUGUUGCG_GGUGCAUCGGAAUGUUGUGGACGGUUUUGU_GGCAAGGAAGGAUCGGGGAGUAAGG ......((.((((((.((....)).)))))).......(..((((..(((((((.(((.(((.....))).)))...)))))))...))))..)..))...................... (-31.26 = -32.06 + 0.80)

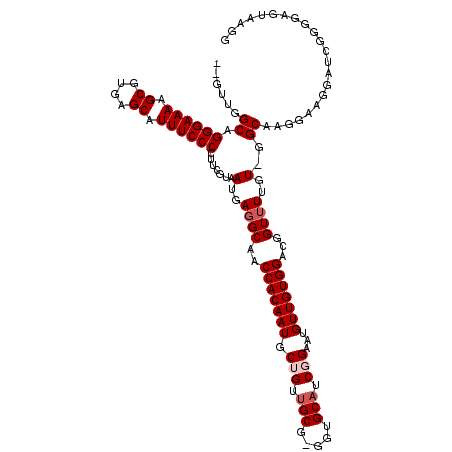
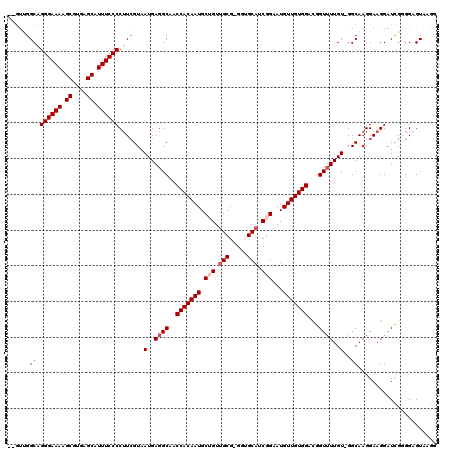
| Location | 14,451,438 – 14,451,551 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.90 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -30.72 |
| Energy contribution | -30.44 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14451438 113 + 27905053 AACAUUCCGAUGCACC-CGCAACAGCAUUGUGGUUGCCUCAUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAAC---UG---CAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAA ..(((((.((.(((.(-(((((.....)))))).))).)).....(..(((((((.((....)).)))))))..)...---..---..((((((......)))))).......))))).. ( -36.80) >DroSec_CAF1 8648 120 + 1 AACAUUCCGAUGCACCCCGCAACAGCAUUGUGGUUGCCUCAUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAACAACUGCAGCAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAA ..(((((.((((((...(((((.....)))))(((((........(..(((((((.((....)).)))))))..).....((((....))))..)))))....))))))....))))).. ( -36.00) >DroSim_CAF1 9242 119 + 1 AACAUUCCGAUGCACC-CGCAACAGCAUUGUGGUUGCCUCAUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAACAACUGCAGCAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAA ..(((((.((.(((.(-(((((.....)))))).))).)).....(..(((((((.((....)).)))))))..).............((((((......)))))).......))))).. ( -36.80) >DroEre_CAF1 8446 113 + 1 AACAUUCCGAAGCCCC-CGCAACUGCAUUGUGGUUGCCUCAUUACUACGGGGAAAUGCUCACGCUUUUCCCGGCCGAC---UG---CAGCAGAGGCAACAUGUUGCAUUUUAUGAAUGAA ..(((((....(((..-.(((((..(...)..)))))...........(((((((.((....))))))))))))....---((---(((((..(....).)))))))......))))).. ( -38.10) >DroYak_CAF1 9757 106 + 1 AACAUUCCGAUGCCCC-CGCAACUGCAUUGUGGUUGCCUCAUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCC-------------GCAGCAGCAACAUGUUGCAUUUUAUGAAUGAA ..(((((.((.((..(-(((((.....))))))..)).)).....(..(((((((.((....)).)))))))..)-------------((((((......)))))).......))))).. ( -34.20) >consensus AACAUUCCGAUGCACC_CGCAACAGCAUUGUGGUUGCCUCAUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAAC___UG___CAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAA ..(((((....((.....(((((..(...)..)))))..........((((((((.((....))))))))))))..............((((((......)))))).......))))).. (-30.72 = -30.44 + -0.28)

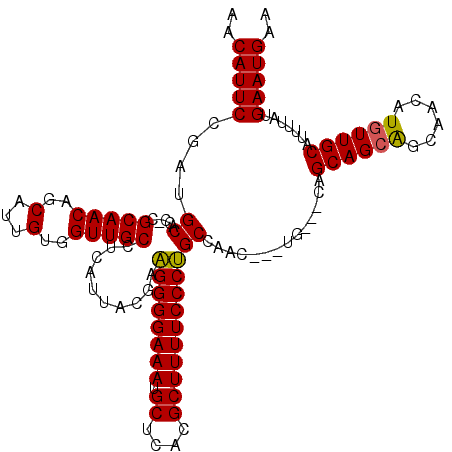
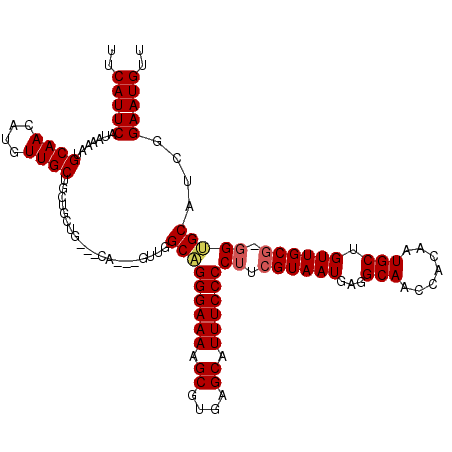
| Location | 14,451,438 – 14,451,551 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.90 |
| Mean single sequence MFE | -40.68 |
| Consensus MFE | -30.68 |
| Energy contribution | -31.04 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14451438 113 - 27905053 UUCAUUCAUAAAAUGCAACAUGUUGCUGCUGCUG---CA---GUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAUGAGGCAACCACAAUGCUGUUGCG-GGUGCAUCGGAAUGUU ..(((((.....(((((...(((((((((....)---))---).)))))((((((.((....)).)))))).((((((((..((((.......))))))))))-)))))))..))))).. ( -38.50) >DroSec_CAF1 8648 120 - 1 UUCAUUCAUAAAAUGCAACAUGUUGCUGCUGCUGCUGCAGUUGUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAUGAGGCAACCACAAUGCUGUUGCGGGGUGCAUCGGAAUGUU ..(((((.....(((((......(((((((((....)))))....))))((((((.((....)).))))))(((((((((..((((.......))))))))))))))))))..))))).. ( -43.20) >DroSim_CAF1 9242 119 - 1 UUCAUUCAUAAAAUGCAACAUGUUGCUGCUGCUGCUGCAGUUGUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAUGAGGCAACCACAAUGCUGUUGCG-GGUGCAUCGGAAUGUU ..(((((.......((((....))))(((..((((.((((((((.((..((((((.((....)).))))))((((.....))))...)))))..))))).)))-)..)))...))))).. ( -40.70) >DroEre_CAF1 8446 113 - 1 UUCAUUCAUAAAAUGCAACAUGUUGCCUCUGCUG---CA---GUCGGCCGGGAAAAGCGUGAGCAUUUCCCCGUAGUAAUGAGGCAACCACAAUGCAGUUGCG-GGGGCUUCGGAAUGUU ..(((((......((((....(((((((((((((---(.---(....).((((((.((....)).)))))).))))))..)))))))).....))))(..((.-...))..).))))).. ( -40.60) >DroYak_CAF1 9757 106 - 1 UUCAUUCAUAAAAUGCAACAUGUUGCUGCUGC-------------GGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAUGAGGCAACCACAAUGCAGUUGCG-GGGGCAUCGGAAUGUU ..(((((.....((((....((((((....))-------------))))((((((.((....)).))))))(((((((((...(((.......))).))))))-)))))))..))))).. ( -40.40) >consensus UUCAUUCAUAAAAUGCAACAUGUUGCUGCUGCUG___CA___GUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAUGAGGCAACCACAAUGCUGUUGCG_GGUGCAUCGGAAUGUU ..(((((.......((((....))))....................(((((((((.((....)).))))))((.((((((...(((.......))).)))))).)))))....))))).. (-30.68 = -31.04 + 0.36)

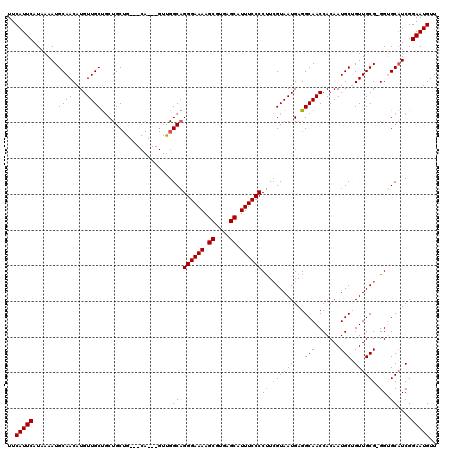
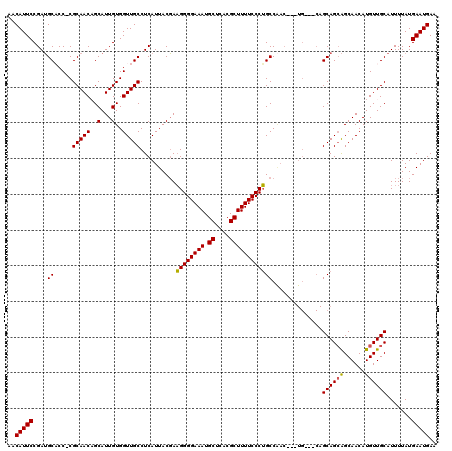
| Location | 14,451,477 – 14,451,591 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -28.20 |
| Energy contribution | -27.92 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14451477 114 + 27905053 AUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAAC---UG---CAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCG .....(..(((((((.((....)).)))))))..)...---.(---(.((((((......))))))..........(.(((...(((((((((.......)))))))))..))).).)). ( -30.40) >DroSec_CAF1 8688 120 + 1 AUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAACAACUGCAGCAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCG .....(..(((((((.((....)).)))))))..)..........((.((((((......))))))..........(.(((...(((((((((.......)))))))))..))).).)). ( -30.60) >DroSim_CAF1 9281 120 + 1 AUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAACAACUGCAGCAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCG .....(..(((((((.((....)).)))))))..)..........((.((((((......))))))..........(.(((...(((((((((.......)))))))))..))).).)). ( -30.60) >DroEre_CAF1 8485 114 + 1 AUUACUACGGGGAAAUGCUCACGCUUUUCCCGGCCGAC---UG---CAGCAGAGGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCG .......((((((((.((....))))))))))((....---((---(((((..(....).))))))).........(.(((...(((((((((.......)))))))))..))).).)). ( -32.40) >DroYak_CAF1 9796 107 + 1 AUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCC-------------GCAGCAGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCG .....(..(((((((.((....)).)))))))..)-------------((.(((((.....)))))..........(.(((...(((((((((.......)))))))))..))).).)). ( -29.10) >consensus AUUACGAAGGGGAAAUGCUCACGCUUUUCCCUGCCAAC___UG___CAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCG .......((((((((.((....))))))))))((..............((((((......))))))..........(.(((...(((((((((.......)))))))))..))).).)). (-28.20 = -27.92 + -0.28)

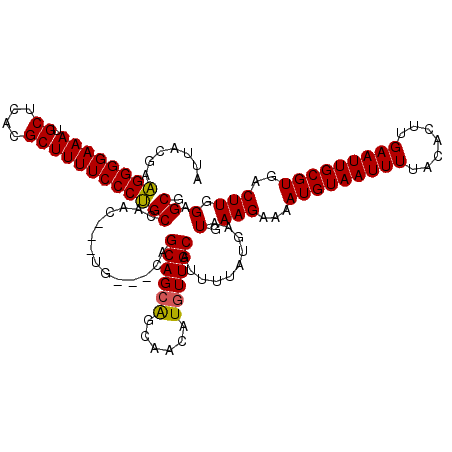
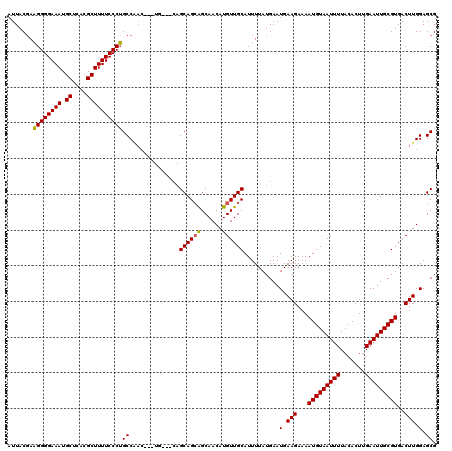
| Location | 14,451,477 – 14,451,591 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.12 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14451477 114 - 27905053 CGCUCCAAGUCACGCAAUUCAAGUGUAAAAUUACAUUUUCUUCAUUCAUAAAAUGCAACAUGUUGCUGCUGCUG---CA---GUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAU ....((((.(((.(((....(((((((....)))))))................((((....))))...)))))---.)---.))))..((((((.((....)).))))))......... ( -27.80) >DroSec_CAF1 8688 120 - 1 CGCUCCAAGUCACGCAAUUCAAGUGUAAAAUUACAUUUUCUUCAUUCAUAAAAUGCAACAUGUUGCUGCUGCUGCUGCAGUUGUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAU ........((((.(((....(((((((....)))))))................((((....)))).(((((....)))))))))))).((((((.((....)).))))))......... ( -31.50) >DroSim_CAF1 9281 120 - 1 CGCUCCAAGUCACGCAAUUCAAGUGUAAAAUUACAUUUUCUUCAUUCAUAAAAUGCAACAUGUUGCUGCUGCUGCUGCAGUUGUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAU ........((((.(((....(((((((....)))))))................((((....)))).(((((....)))))))))))).((((((.((....)).))))))......... ( -31.50) >DroEre_CAF1 8485 114 - 1 CGCUCCAAGUCACGCAAUUCAAGUGUAAAAUUACAUUUUCUUCAUUCAUAAAAUGCAACAUGUUGCCUCUGCUG---CA---GUCGGCCGGGAAAAGCGUGAGCAUUUCCCCGUAGUAAU .(((.(..((((((((....(((((((....)))))))................((((....))))......))---).---)).))).((((((.((....)).)))))).).)))... ( -26.80) >DroYak_CAF1 9796 107 - 1 CGCUCCAAGUCACGCAAUUCAAGUGUAAAAUUACAUUUUCUUCAUUCAUAAAAUGCAACAUGUUGCUGCUGC-------------GGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAU .............(((....(((((((....)))))))................((((....)))))))(((-------------((..((((((.((....)).))))))..))))).. ( -28.30) >consensus CGCUCCAAGUCACGCAAUUCAAGUGUAAAAUUACAUUUUCUUCAUUCAUAAAAUGCAACAUGUUGCUGCUGCUG___CA___GUUGGCAGGGAAAAGCGUGAGCAUUUCCCCUUCGUAAU ........(((.........(((((((....)))))))................((((....))))...................))).((((((.((....)).))))))......... (-23.12 = -23.12 + -0.00)

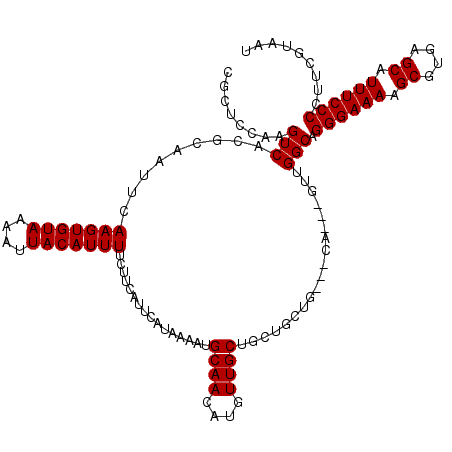
| Location | 14,451,515 – 14,451,631 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.57 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14451515 116 + 27905053 -UG---CAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCGGUGAGUUCUCAUAUUUCAUAGCCGCUCAAAGAUGCCGCAA -((---(.((((((((.....)))))..................(((((((((.......)))))))))..(((.(((((((((((......))))....))))))).))).))).))). ( -32.80) >DroSec_CAF1 8728 120 + 1 CUGCAGCAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCGGUGAGUUCUCAUAUUUCAUAGCCGCUCAAAGAUGCCGCAA .(((.(((((((((......))))))..................(((((((((.......)))))))))..(((.(((((((((((......))))....))))))).))).))).))). ( -34.50) >DroSim_CAF1 9321 120 + 1 CUGCAGCAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCGGUGAGUUCUCAUAUUUCAUAGCCGCUGAAAGAUGCCGCAA .(((.(((.((((.((((....))))...(((((((..(((...(((((((((.......)))))))))..)))(((((.....))))).....)))))))..)))).....))).))). ( -33.10) >DroEre_CAF1 8523 112 + 1 -UG---CAGCAGAGGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCGGUAAGUUCUCAUAUUUCAUAGCC----CAAGAUGCCGCAA -((---(.(((..(((..((((........)))).((((((...(((((((((.......))))))))).....(((((.....)))))....)))))).)))----.....))).))). ( -26.40) >DroYak_CAF1 9831 108 + 1 --------GCAGCAGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCGGUGGGUUCUCAUAUUUCAUAGCC----CAAGAUGCCGCAA --------((.(((((((....))))..................(((((((((.......)))))))))..(((((.((.(((((.........))))).)))----)))).))).)).. ( -27.90) >consensus _UG___CAGCAGCAGCAACAUGUUGCAUUUUAUGAAUGAAGAAAAUGUAAUUUUACACUUGAAUUGCGUGACUUGGAGCGGUGAGUUCUCAUAUUUCAUAGCCGCU_AAAGAUGCCGCAA ........((.(((((((....))))...(((((((..(((...(((((((((.......)))))))))..)))(((((.....))))).....)))))))...........))).)).. (-24.66 = -24.70 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:50 2006